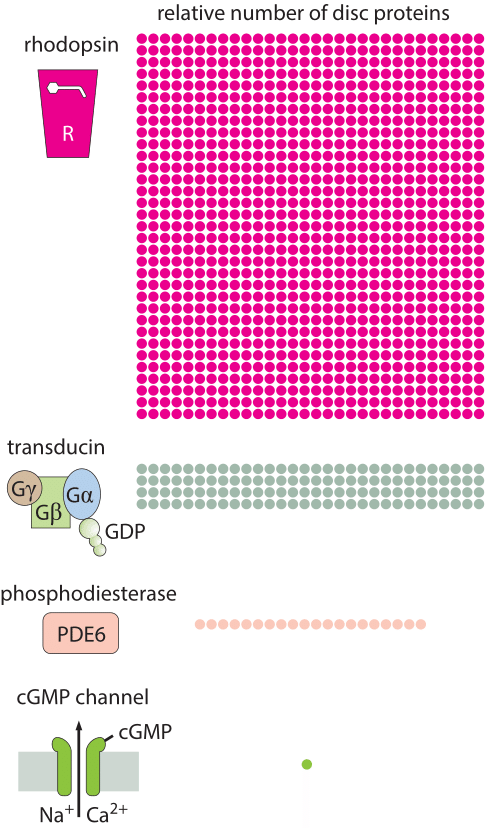
How many rhodopsin molecules are in a rod cell?
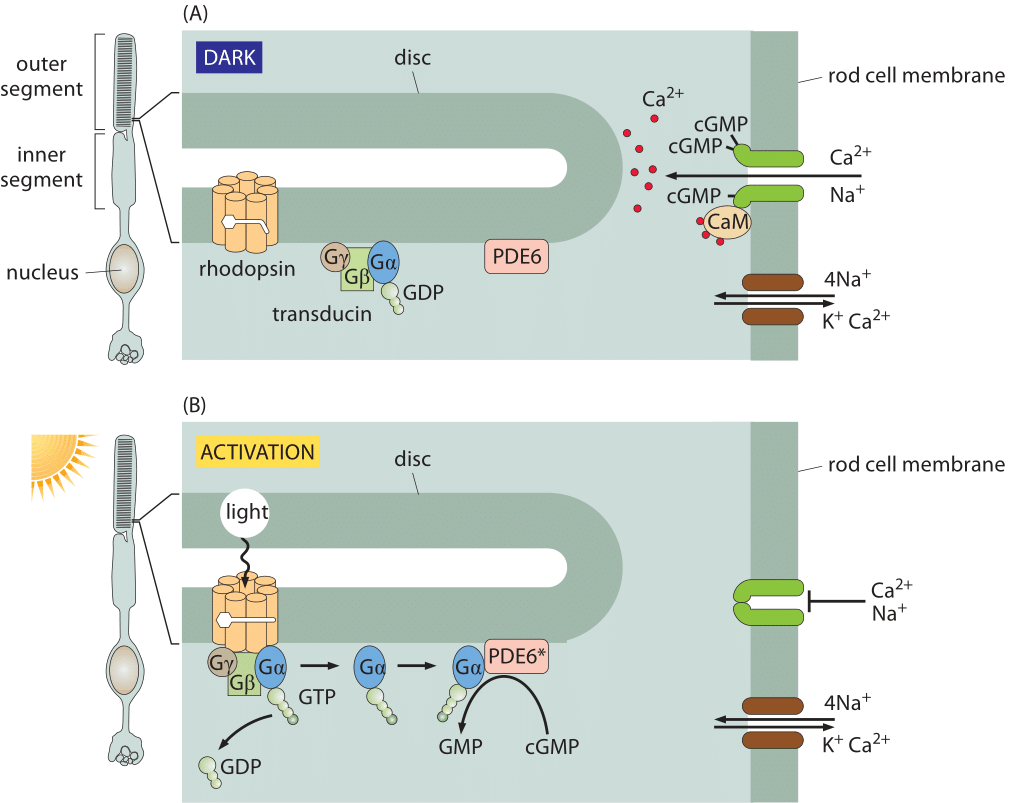
Figure 1: Signal transduction in the retina. (A) In the dark, the rhodopsin is in the inactive state and ions are free to cross the rod cell membrane. In the dark, the cGMP phosphodiesterase PDE6 is inactive, and cGMP is able to accumulate inside the rod cell. cGMP binds to a ligand-gated ion channel (dark green) that is permeable to both sodium and calcium ions. Calcium is transported back out again by an exchanger (shown in brown) that uses the energy from allowing sodium and potassium ions to run down their electrochemical gradients to force calcium ions to be transported against their gradient. (B) Activation of rhodopsin by light results in the hydrolysis of cGMP, causing cation channels to close. When a photon activates a rhodopsin protein, this triggers GTP-for-GDP exchange on transducin, and the activated α subunit of transducin then activates PDE6, which cleaves cGMP. The ligand-gated channels close, and the transmembrane potential becomes more negative (adapted from A. Stockman et al., Journal of Vision 8: 1, 2008.)
Responses in signaling pathways depend critically upon how many molecules there are to respond to the signal of interest. The concentrations of molecules such as rhodopsin in photoreceptor cells determine the light intensity that can be detected in vertebrate eyes. Beyond this, the number of rhodopsins also helps us understand how frequently a given rod cell will spontaneously fire in the dark. Though our focus on rhodopsin might seem highly specialized, we find it an informative case study as the signaling cascade associated with vision is one of the best characterized of human signaling cascades. Further, it exhibits many generic features found in signaling events of many other kinds. Some of the key molecular players found here include G-coupled receptors and ligand-gated ion channels, molecules in signaling cascades that are ubiquitous throughout the living world. Figure 1 shows how the molecules in the outer segment of a photoreceptor respond to the arrival of a photon which is absorbed by the retinal pigment, covalently but reversibly held by the opsin protein, together making up the rhodopsin molecule.
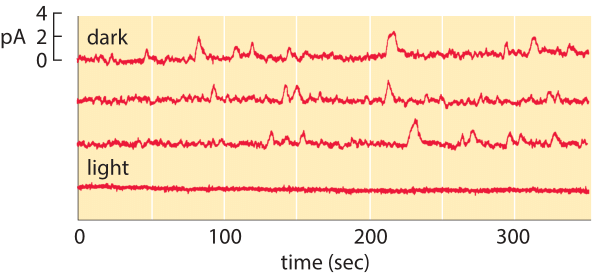
Figure 2: Spontaneous isomerization of retinal. The top three traces correspond to the current measured from a single photoreceptor as a function of time in the dark. The lower trace shows the current in the light and demonstrates that the channels are closed in the presence of light. (Adapted from D. A. Baylor et al., J. Physiol. 309:591, 1980.)
In this vignette, we use a collection of estimates to work out the number of rhodopsins in a photoreceptor cell. We begin by estimating the number of membrane discs in the outer segment of a rod cell. As seen in both the electron microscopy image and associated schematic in the vignette on “How big is a photoreceptor?”, the rod outer segment is roughly 25 µm in length and is populated by membrane discs that are roughly 10 nm thick and 25 nm apart. This means there are roughly 1000 such discs per rod outer segment. Given that the rod cell itself has a radius of around 1 µm, this means that the surface area per disc is roughly 6 µm2, resulting in an overall membrane disc area of 6000 µm2. One crude way to estimate the number of rhodopsins in each rod cell outer segment is to make a guess for the areal density of rhodopsins in the disc membranes. Rhodopsins are known to be tightly packed in the disc membranes and we can estimate their mean spacing as 5-10 nm (i.e. about one to two diameters of a characteristic protein), corresponding to an areal density of s= 1/25 – 1/100 nm-2. In light of these areal densities, we estimate the number of rhodopsins per membrane disk to be between (6 x 106 nm2) x (1/25 nm-2) ≈ 2 x 105 and (6 x 106 nm2) x (1/100 nm-2) = 6 x 104. The actual reported numbers are ≈105 rhodopsins per membrane disc or ≈108 per photoreceptor (BNID 108323), which is on the order of the total number of proteins expected for such cell volume as discussed in the vignette on “How many proteins are in a cell?”. This tight packing is what enables the eye to be able to function so well at extremely low light levels.
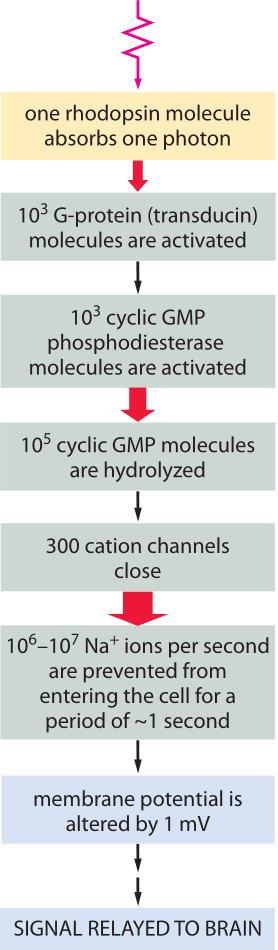
Figure 3: Signal amplification is achieved at several steps of the pathway, such that the energy of one photon eventually triggers a net charge change of about one million sodium ions.
For a molecule such as retinal that absorbs photons, it is convenient to define the effective cross section which quantifies its absorption capacity. Concretely, the absorption cross section is defined as that area perpendicular to the incident radiation such that the photon flux times that area is equal to the number of photons absorbed by the molecule. The cross section for absorption of retinal is (absorption ability) with the geometrical notion of area (of the photoreceptor). Here is an example of why this is useful. With 108 rhodopsins per photoreceptor we arrive at a total absorption cross section of (108 rhodopsin/photoreceptor) x (10-2 nm2/rhodopsin) = 106 nm2/photoreceptor = 1 µm2. Each photoreceptor cell has a geometrical cross sectional area of about 4 µm2/photoreceptor as discussed in the vignette on “How big is a photoreceptor?”. From the similarity between the cross section for absorption and the actual cross sectional area of the photoreceptor we can infer that the concentration of rhodopsins is of the correct order of magnitude to efficiently absorb all photons arriving (at least under low illumination levels when reactivation of rhodopsin following absorption does not become limiting). This does not mean there are no lost photons. Indeed, to achieve superior night vision, many nocturnal animals have a special layer under the retina called the tapetum lucidum that acts as a reflector to return the photons that were not absorbed by the rhodopsins back to the retina for another opportunity to be absorbed. This increases their ability to hunt prey at night and enables naturalists to find hyena, wolves (and also domestic cats) at night from a distance, by looking for eyeshine, which is the reflection when pointing a flashlight.
The membrane census of rhodopsin also sheds light on the rate at which rod cells suffer spontaneous thermal isomerizations of the pigment retinal. As shown in Figure 2, measurements of the currents from individual rod photoreceptors exhibit spontaneous isomerizations. Reading off of the graph, we estimate roughly 30 spontaneous events over a period of 1000 seconds corresponding to a rate of once per 30 seconds. We know that the total rate is given by (rate)= (# rhodopsins) x (rate/rhodopsin). Armed with the number of rhodopsins estimated above, we deduce that the rate of spontaneous isomerization per rhodopsin is once about every 100 years.
The signaling cascade that follows the absorption of a photon only starts with the isomerization of a retinal molecule. Once the rhodopsin molecule has been thus activated, it sets off a signaling cascade within the rod cell that amplifies the original signal as already shown in Figure 1 and elaborated on more quantitatively in Figure 3. In particular, once the rhodopsin has been activated, it encounters a membrane bound G-protein coupled receptor and activates its alpha subunit. Over a period of 100 ms, one activated rhodopsin will create ≈103 of these activated alpha subunits (Ga) as explained in detail in the wonderful book “The first steps in seeing” by R. W Rodieck. These molecules bind another molecule known as phosphodiesterase which can convert cyclic guanosine monophosphate into guanosine monophosphate. The significance of this molecular reaction is that it is the cyclic guanosine monophosphate that gates the cGMP channels in the rod cell membrane that lead to the change in membrane potential upon excitation. Hence, the activation of the receptor via photons leads to a closing of the channels and a change in the membrane potential. The census of the various molecular players in this signaling cascade is shown in Figure 3. Though we can depict the molecular details and the associated copy numbers as an advanced Rube Goldberg machine, the fitness advantage of this specific design beyond the obvious need to amplify a small signal, is still quite a mystery even to researchers in the field. As a parting note we consider another amazing number related to the function of the rod cell. Every pigment molecule, once it absorbs a photon, is photobleached and it takes about 10 minutes and a sequence of biochemical steps after transport to a separate organelle to fully regenerate (BNID 111399, 111394). The inventory of pigments in the rod cell have to compensate for this long delay.