How big are nuclei?
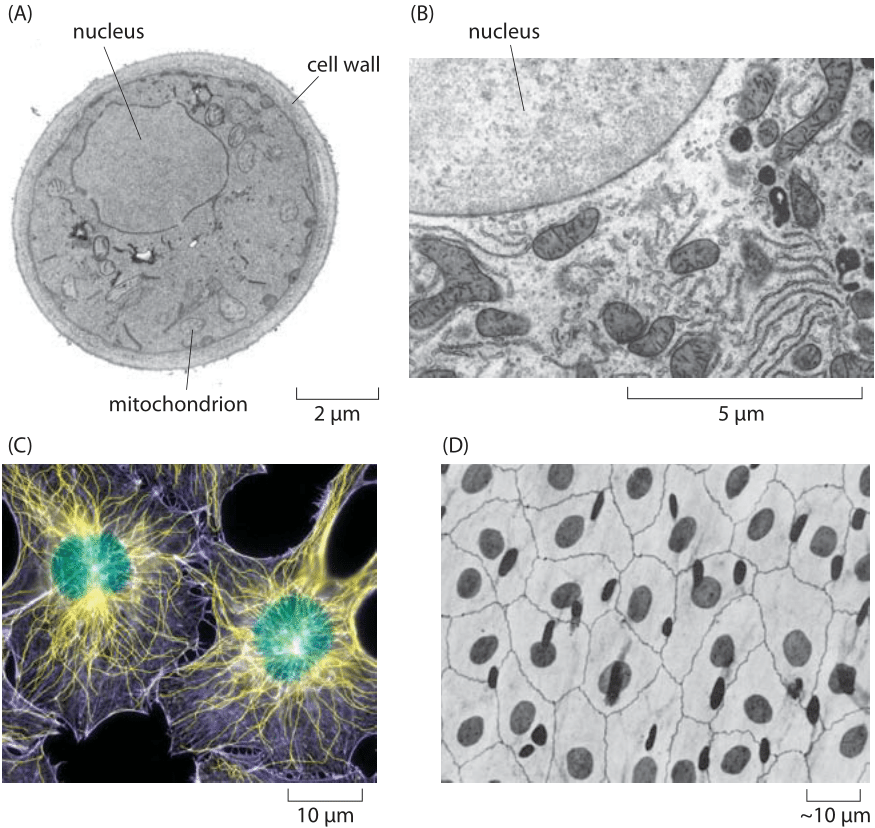
Figure 1. Nuclear size. (A) Electron microscopy image of a yeast cell revealing the roughly 2 micron-sized nucleus. (B) A portion of a rat liver cell showing part of the nucleus and a variety of surrounding organelles such as the endoplasmic reticulum, mitochondria and the Golgi apparatus. (C) Fluorescence image of a human fibroblast cell with the roughly 10 micron nucleus labeled in green. (D) Light microscopy image of a human epithelial sheet. The dark ovals are the cell nuclei stained with silver. (B, adapted from electron micrograph from D. W. Fawcett, The Cell, Its Organelles and lnclusions: An Atlas of Fine Structure Philadelphia, W. B. Saunders & Co., 1966., A, MBOC, C, D, PBOC)
One of the most intriguing structural features of eukaryotic cells is that they are separated into many distinct compartments, each characterized by differences in molecular composition, ionic concentrations, membrane potential and pH. In particular, these compartments are separated from each other and the surrounding medium (i.e. cytoplasm or extracellular solution) by membranes which themselves exhibit a great diversity of lipid and protein molecules, with the membranes of different compartments also characterized by different molecular compositions. Given the central role of genomes in living matter, there are few organelles as important as the eukaryotic nucleus, home to the chromosomal DNA that distinguishes one organism from the next. As seen in Figure 1, using both electron and light microscopy it is possible to determine nuclear size variation with typical diameters ranging between 2 and 10 microns, though in exceptional cases such as oocytes, the nuclear dimensions are substantially larger.
One feature of organellar dimensions is their variability. We have already seen the range of sizes exhibited by yeast cells in an earlier vignette. Figure 2 takes up this issue again by revealing the typical sizes and variability for the nuclei in haploid and diploid yeast cells, complementing the data presented earlier on cell size. For haploid yeast cells, the mean nuclear volume is 3 μm3 (BNID 104709). With a genome length of 12 Mbp (BNID 100459), the DNA takes up roughly 0.3 % of the nuclear volume. We can arrive at this estimate based on the rule of thumb that a base pair has a volume of ≈1 nm3 (BNID 103778)) and thus the DNA occupies roughly 0.01 μm3. A similar value is found for diploid yeast. In contrast, for the yeast spore, the nuclear volume is an order of magnitude smaller – 0.3 μm3 (BNID 107660) or about 3% of the nuclear volume – indicating a much more dense packing of the genomic DNA.
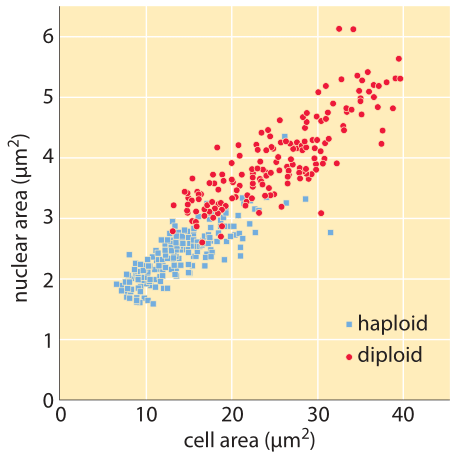
Figure 2: Nuclear size for haploid and diploid yeast cells. The cross sectional area of the nuclei are plotted as a function of the cross sectional area of the cells themselves. (Adapted from P. Jorgensen et al., Molecular Biology of the Cell, 18:3523, 2007. )
These estimates for nuclear fraction are agnostic of the higher-level chromatin structure induced by nucleosome formation. In nucleosomes, 147 base pairs of DNA are wrapped roughly 1¾ times around an octamer of histone proteins making a snub disk roughly 10 nm across (BNID 102979, 102985). In Figure 3 we show the so-called 30 nm fiber. When we travel 10 nm along the fiber, about 6 nucleosomes are packed in a staggered manner, and thus we have included on the order of 1000 bp. We can estimate the total volume taken up by the genomic DNA of yeast when in this structure by multiplying the area of the effective circular cross section by the height of the structure resulting in V=p (15 nm)2 x (10nm/1000 bp) x (107 bp) ≈ 108 nm3 = 0.1 μm3. Given the volume of the yeast nucleus of roughly 4 mm3, this implies a packing fraction of ≈2%, and is consistent with our earlier estimate which was based on the volume of a base pair.
Questions about nuclear size in eukaryotes have been systematically investigated in other organisms besides yeast. It has been hypothesized that there is a simple linear relationship between the mean diameter of a plant meristematic cell (the plant tissue consisting of undifferentiated cells from which growth takes place) and the diameter of its nucleus. Such ideas have been tested in a variety of different plant cells, as shown in Figure 4, for example. In the experiments summarized there, the nuclear and cell volumes of 14 distinct species of herbaceous angiosperms including some commonly known plants such as chickpeas and lily of the valley were measured, resulting in a simple relationship of the form Vnuc ≈ 0.2 Vcell (BNID 107802).
The observations reported here raise the question of how the relative size of the nucleus to the whole cell is controlled. This is especially compelling since the nucleus undergoes massive rearrangements during each and every cell cycle as the chromosomes are separated into the daughter cells. We remind in ending that a relatively stable ratio is a common observation rather than a general law. In mammalian cells this ratio can be very different between cell types. For example, in resting lymphocytes the nucleus occupies almost the whole cell while in macrophages or fat cells, the ratio of nucleus to cell volume is much smaller.
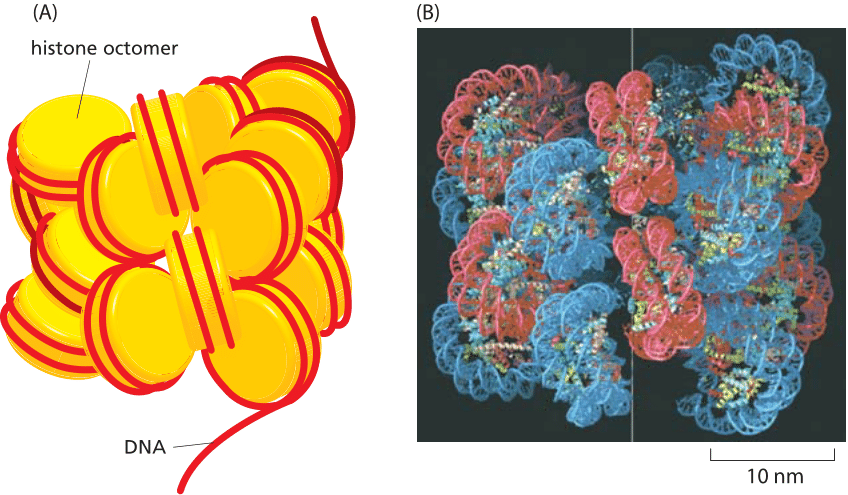
Figure 3: DNA packing into higher-level compact structures. (A) Schematic illustrating how multiple nucleosomes can be arranged into a solenoidal structure. Histone octamer shown in yellow and DNA as red strand. (B) Models of nucleosome packing based upon high-resolution cryo-electron microscopy images of arrays of nucleosomes. In these in vitro experiments, nucleosome arrays were generated by using purified histones and specific DNA molecules of known sequence.(B adapted from P. Robinson, L. Fairall, V. Huynh and D. Rhodes, Proc. Natl Acad. Sci. U.S.A. 103:6506-6511, 2006.)

Figure 4: The relationship between the nuclear volume and cell volume in apical meristems of 14 herbaceous angiosperms. 1 Arabidopsis thaliana, 2 Lobularia maritime (Sweet Alison), 3 Hypericum virginicum (Marsh St. John’s wort), 4 Cicer arietinum (chickpea), 5 Nelumbo lutea, 6 Spinacia oleracea (spinach), 7 Cyanotis pilosa, 8 Anemone pulsatilla (Meadow Anemone), 9 Tradescania navicularis (day flower), 10 Convallaria majalis (Lily of the valley), 11 Fritillaria laneeolata (chocolate lily), 12 Fritillaria camtschatcensis, 13 Lilium longiflorum (Easter lily)(4 x ), 14 Sprekelia formosissima (Aztec lily). (Adapted from H. J. Price et al., Experientia, 29:1028, 1973.)

