How many mRNAs are in a cell?
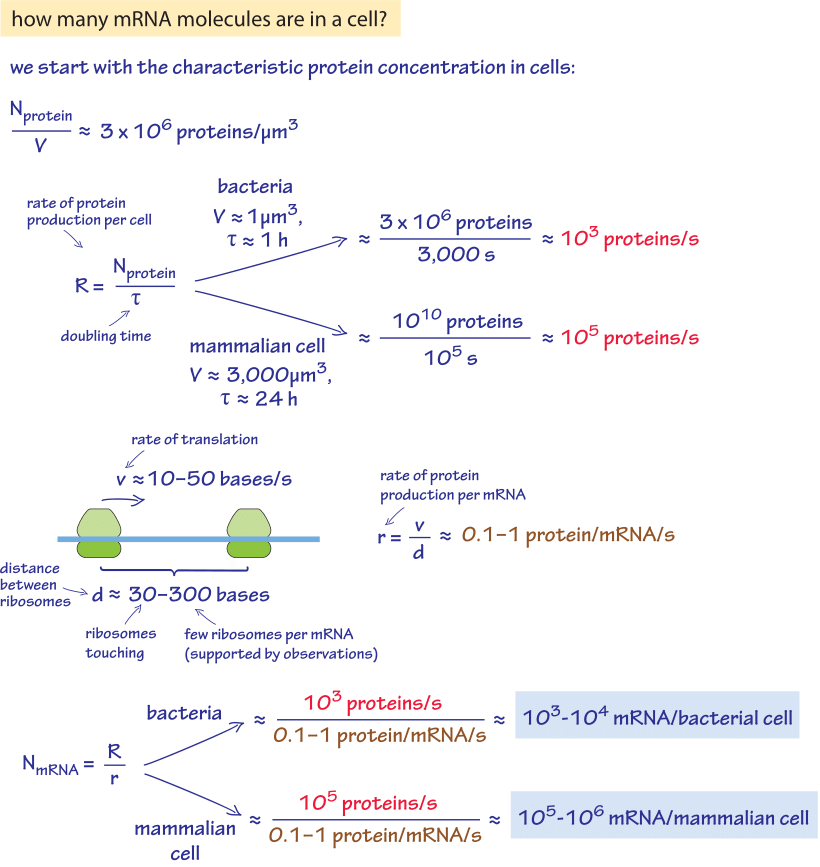
Figure 1: A back of the envelope calculation of the number of proteins per cell in a characteristic bacteria and mammalian cell. Estimate is based on generic parameter values. For more accurate organism specific values see main text.
Given the central place of gene regulation in all domains of biology, there is great interest in determining the census of mRNA in cells of various types. We are interested both in specific genes and in the entire transcriptome as a function of environmental conditions and developmental stage. Such measurements provide a direct readout of the instantaneous regulatory state of the cell at a given time and as such, give us a powerful tool to analyze how cellular decision making is implemented. We begin with an exercise of the imagination to see if we can use a few key cellular facts in order to estimate the number of mRNAs. Given our knowledge of the kinetics of the processes of the central dogma during one cell cycle, the number of mRNA molecules per cell can be worked out as shown in Figure 1. The essence of the estimate is to exploit the recognition that over the course of the cell cycle, the number of proteins must be doubled through protein synthesis. This protein synthesis is based, in turn, on the distribution of mRNA molecules that are present in the cell. As shown in this back of the envelope calculation we can derive an estimate for rapidly dividing cells of 103-104 mRNA per bacterial cell and 105-106 mRNA per the 3000 μm3 characteristic size of a mammalian cell.
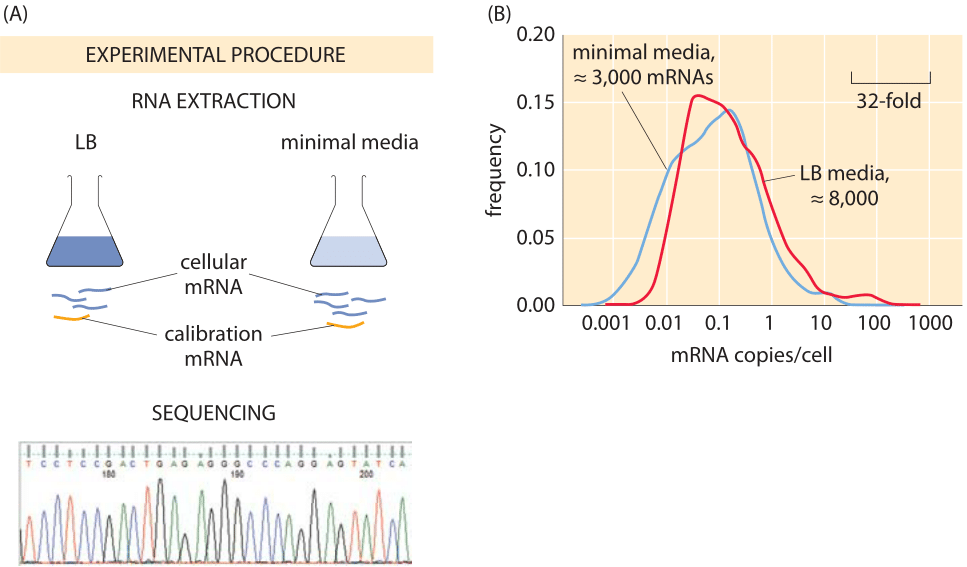
Figure 2: Using sequencing to find the number of mRNAs per cell. (A) mRNA is carefully extracted from cells and mixed with synthesized mRNA that serves for calibration. Deep sequencing enables counting the number of copies of each mRNA type and from this the total number of mRNAs can be inferred. (B) mRNA counts for E. coli grown in rich media and minimal media. (Data courtesy of Zoya Ignatova)
Modern techniques have now largely superseded those leading to the classic census numbers. One approach is oriented towards “genome-wide” measurements in which an attempt is made to size up the number of mRNAs across the entire transcriptome. These results are based upon the method of RNA-Seq where individual mRNAs are sequenced from the cell lysate. Of course, since the count is based on the frequency of sequence reads, it requires calibration. To that end, the sample is spiked with mRNAs standards whose quantity is known prior to sequencing. An example of such a result is shown in Figure 2, which reports on the distribution of mRNAs in E. coli grown under both rich and minimal media conditions. The result of this study is that the number of transcripts per cell (≈8000 mRNA copies/cell) for cells grown in LB media is roughly three-fold larger than the number of transcripts per cell (≈3000 mRNA copies/cell) for cells grown in minimal media. Given that the number of genes is in excess of 4000, this implies that the mean copy number in LB is on the order of one per cell. For most genes it is actually even less, meaning that in most cells there are zero copies while in some there is one or two. We find this striking fact to be one example of where for most people the picture of the contents of a cell, in this case a bacterial cell, gets augmented through the usage of numbers as a sixth sense to perceive cells.
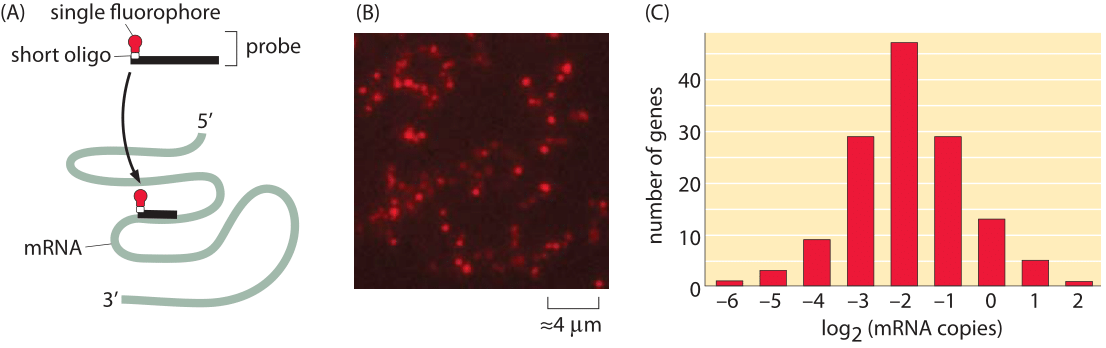
Figure 3: Fluorescence microscopy approach to taking the mRNA census. Using fluorescence in-situ hybridization (FISH), it is possible to use fluorescence intensity of specific probes that hybridize to an mRNA of interest to count these mRNA. (A) Schematic of the single-molecule probes used to label mRNA. (B) Fluorescence image of a field of E. coli cells with the mRNA for a specific gene imaged. (C) Histogram of the mean number of mRNA in E. coli for a number of genes. (Adapted from Taniguchi et al. Science, 329, 533 (2010)).
One of the most important questions at the center of the biological numeracy called for in our book is that of reproducibility, especially when different methods are brought to bear on the same problem. A very useful alternative for taking the mRNA census is built around direct counting by looking at the individual mRNA molecules under a microscope. Specifically, a gene-by-gene decomposition using techniques such as single-molecule fluorescence in-situ hybridization (FISH) complements the RNA-Seq perspective described above. FISH provides a window onto the mRNA spatial and cell-to-cell distribution for a particular species of mRNA molecule as shown here in Figure 3 and as shown in Figure 2 of the vignette on “How much cell-to-cell variability exists in protein expression?”. The idea in this case is to design probes that bind to the mRNA of interest through complementary base pairing. Each such probe harbors a fluorophore and hence, when the fixed cells are examined in the microscope, the intensity of the fluorescence of these probes is used as a readout of the number of mRNAs. As seen in Figure 3B, the number of mRNAs per cell is generally between 0.1 and 1, with several outliers having both smaller and larger mRNA counts. As has also been emphasized throughout the book, different conditions result in different numbers and the FISH results recreated here correspond to slow growth.
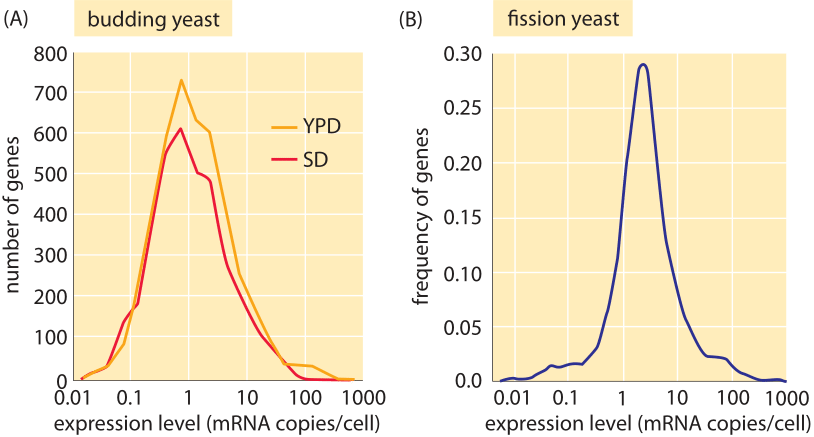
Figure 4: mRNA distributions in yeast. (A) mRNA distribution in budding yeast grown in rich media (YPD) and minimal media (SD) as measured using PCR. (B) mRNA distribution in fission yeast measured using RNA-Seq. ((A) adapted from F. Miura, BMC Genomics, 9, 574 (2008); (B) adapted from S. Marguerat, Cell, 151:671 (2012)).
What about the mRNA census in other cell types besides bacteria? Again, both sequencing methods and microscopy have been brought to bear on these questions in yeast and other eukaryotes. Figure 4 presents results for the mRNA census in both budding and fission yeast. The total number of mRNA per cell is in the range 20,000-60,000 in exponentially growing budding yeast (BNID 104312, 102988, 103023, 106226, 106763). As with our earlier results for bacteria, here too we find that each gene generally only has a few mRNA molecules present in the cell at any one time. The vignette on “What is the protein to mRNA ratio in cells?” provides a window onto the amplification factor that attends a given mRNA as it is turned into the proteins of the cell in the process of translation. For “typical” mammalian cells a quoted value of 200,000 mRNA per cell (BNID 109916) is in line with our simple estimate above and shows that scaling the number of mRNA proportionally with size and growth rate seems to be a useful first guess.

