How big are the molecular machines of the central dogma?
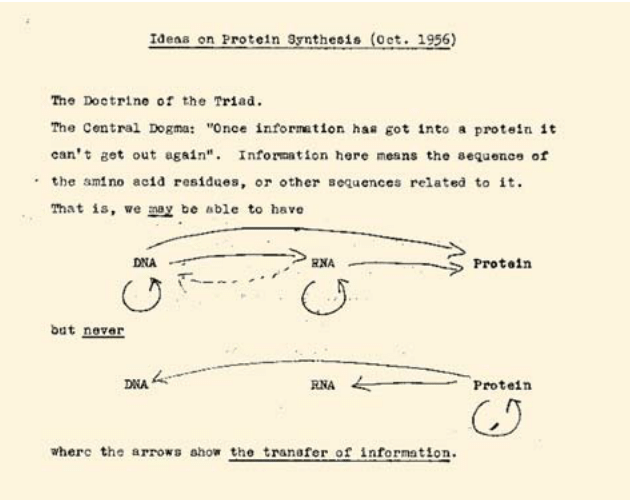
Figure 1: Notes of Francis Crick on the central dogma. Early draft for article published as: Crick, F.H.C. (1958): On Protein Synthesis. Symp. Soc. Exp. Biol. XII, 139-163. The 1958 paper did not include this visual depiction which later appeared in a 1970 Nature paper.
Molecular machines manage the journey from genomic information in DNA to active and functioning protein in the processes of the central dogma. The idea of directional transfer of information through a linked series of processes, termed the central dogma, started out as a fertile hypothesis in the hands of Francis Crick as shown in Figure 1 dated to 1956. In the time since its original suggestion, this hypothesis has been confirmed in exquisite detail, with the molecular anatomy of the machines that carry out these processes now coming into full relief.
The machines that mediate the processes of the central dogma include RNA polymerase, which is the machine that takes the information stored in DNA and puts it in a form suitable for protein synthesis by constructing messenger RNA molecules, and the ribosome, the universal translation machine which synthesizes proteins. Of course, proteins do not survive indefinitely and their fate is often determined by another molecular machine, the proteasome – the central disposal site that degrades the proteins so carefully assembled by the ribosome. Our understanding of these macromolecular complexes has evolved from the point where three to four decades ago, it was only possible to infer their existence, to the present era in which it is possible to acquire atomic resolution images of their structures in different conformational states.
As seen in Figure 1, there is an arrow from DNA to itself which signifies DNA replication. This process of replication is carried out by a macromolecular complex known as the replisome. The E. coli replisome is a collection of distinct protein machines that include helicase (52 kDa (each of 6 subunits) BNID 104931), primase (65 kDa BNID 104932) and the DNA polymerase enzyme complex (791 kDa in several units of the complex, BNID 104931). To put the remarkable action of this machine in focus, an analogy has been suggested in which one thinks of the DNA molecule in human terms by imagining it to have a diameter of 1 m (T. A. Baker & S. P. Bell, Cell 92:295, 1998, to get a sense of the actual size of the replication complex relative to its DNA substrate, see Figure 2). At this scale, the replisome has the size of a FedEx truck, and it travels along the DNA at roughly 600 km/hr. Genome replication is a 400 km journey in which a delivery error occurs only once every several hundred kilometers, this despite the fact that a delivery is being made roughly six times for every meter traveled. During the real replication process, the error rate is even lower as a result of accessory quality control steps (proofreading and mismatch correction) that ensure that a wrong delivery happens only once in about 100 trips.
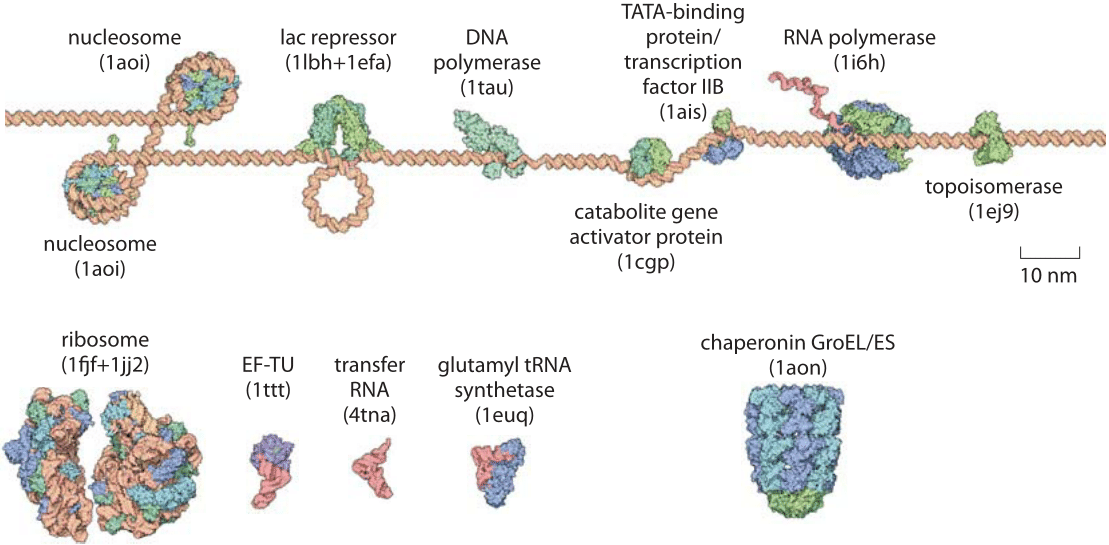
Figure 2: Structures of the machines of the central dogma. The machines responsible for replication, transcription and translation are all shown drawn to scale relative to the DNA substrate. The notations in parenthesis are the PDB database names for the protein structures shown.
Transcription is another key process in the Central Dogma and is intimately tied to the ability of cells to “make decisions” about which genes should be expressed and which should not at a given place within an organism at a given moment in time. The basal transcription apparatus is an assembly of a variety of factors surrounding the RNA polymerase holoenzyme. As shown in Figure 2, the core transcription machinery, like many oligomeric proteins, has a characteristic size of roughly 5 nm and a mass in E. coli of roughly 400 kDa (BNID 104927, 104925). Comparison of the machines of the central dogma between different organisms has been the most powerful example of what Linus Pauling referred to as using “molecules as documents of evolutionary history”. Polymerases have served in that capacity and as such the prokaryotic and eukaryotic polymerases are contrasted in Figure 3.
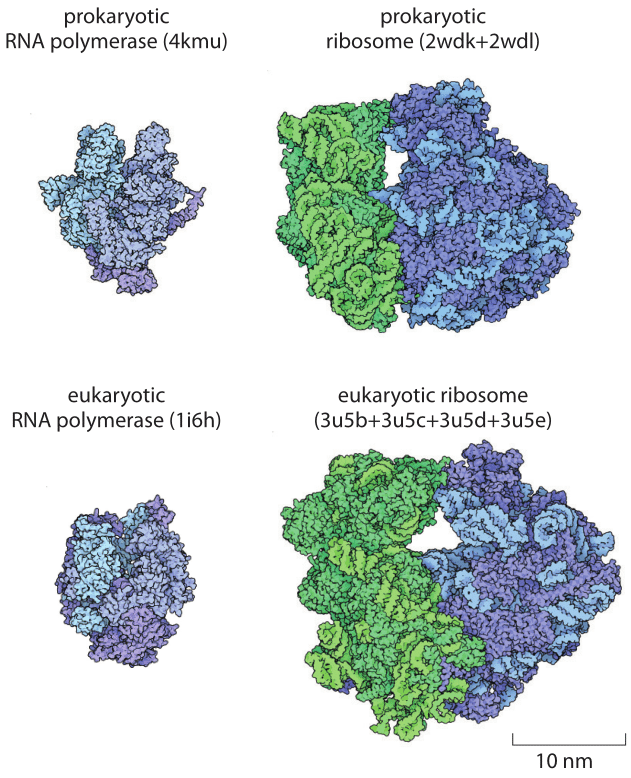
Figure 3: Comparison of the structures of the RNA polymerase and ribosomes from prokaryotic and eukaryotic (in this case yeast) organisms. The yeast ribosome at 3.3 MDa is intermediate between the bacterial ribosome at about 2.5 MDa and the mammalian ribosome at 4.2 MDa (BNID 106865). The notations in parenthesis are the PDB database names for the protein structures shown.
The ribosome, a collection of three RNA chains (BNID 100112) and over 50 proteins (56 in bacteria, BNID 100111 and 78-79 in eukaryotes http://tinyurl.com/l7yykj), is arguably the most studied of all of the machines of the central dogma. Its importance can be seen from any of a number of different perspectives. For fast growing microorganisms like E. coli it can make up over a third of the total protein inventory. From a biomedical perspective it is the main point of attack of many of the most common and effective antibiotics that utilize the intricate differences between the bacterial and eukaryotic ribosomes to specifically stop translation of the former and halt their growth. The ribosome has also served as the basis of a quiet revolution in biology that has entirely rewritten the tree of life. Because of its universality, the comparison of ribosomal sequences from different organisms has served as the basis of a modern version of phylogeny which tells a story of the history of life like no other.
Befitting its central role, the ribosome is also a relatively large molecular machine with a diameter of 20-30 nm (BNID 102320, 111542). In E. coli it is composed of ≈7500 amino acids (BNID 101175, 110217, 110218) and ≈4,600 nucleotides (BNID 101439) with a total mass of 2.5 MDa (BNID 106864, 100118, if it was made only of carbon atoms there will be about 200,000 of them). Given that the characteristic mass of an amino acid is ≈100 Da (BNID 104877) and that of an RNA nucleotide ≈300 Da (BNID 104886), these numbers imply that the RNA makes up close to 2/3 of the mass of the ribosome and proteins only a third. Indeed, crystal structures have made it clear that the function of the ribosome is performed mainly by the RNA fraction, exposing its origins as a ribozyme, an enzyme based on catalytic RNA. The ribosome volume is ≈3000-4000 nm3 (BNID 111543, 104919, 102473, 102474), implying that for rapidly dividing cells a large fraction of the cellular volume is taken up by ribosomes, a truth that is now seen routinely in cryo electron microscopy images of bacteria.
Ending with a somewhat less dogmatic view of the central dogma, the diligent reader might have noticed the broken line in Crick’s note from RNA back to DNA. This feat is achieved through reverse transcriptase which in HIV is a heterodimer of 70 and 50 kDa subunits with a DNA polymerization rate of 10-100 nuc/s (BNID 110136, 110137).

