How fast do molecular motors move on cytoskeletal filaments?
Molecular motors are central to a vast array of different processes with examples including cell crawling, cell division, chromosome segregation, intracellular trafficking, etc. These active processes are driven by motors of many different types moving about on both actin and microtubule filaments. We noted in the vignette on “What are the time scales for diffusion in cells?” (i.e. those bigger than several tens of microns) diffusion times become exorbitant and cells need to resort to motor-mediated directed transport, paid for at the cost of ATP hydrolysis. The presence of these motors makes it possible for cargos of many types including transport vesicles and even organelles to be directed to various places throughout the cell.
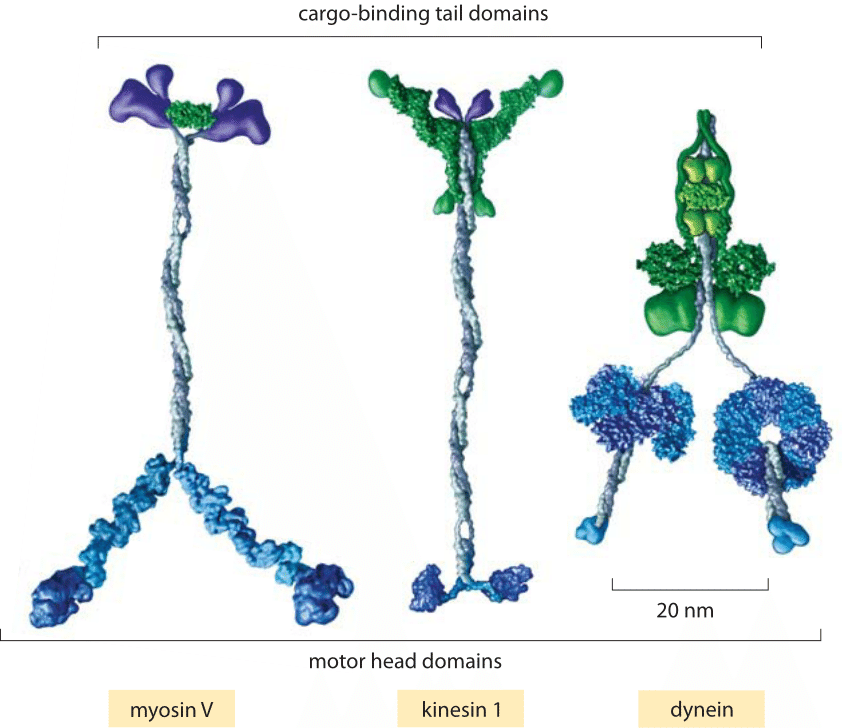
Fig. 1: Key classes of translational motor. (A) A myosin V molecule is one of about 20 different types of myosins that move on actin filaments. (B) Kinesin 1 is also a member of a large family of related molecular motor proteins, but these move on microtubules rather than on actin. Although myosins and kinesins have different substrates, the detailed structures of their motor heads are quite similar and they are thought to be derived from a single common molecular ancestor. (C) Cytoplasmic dynein represents a different class of microtubule-based motors that appears to be unrelated to kinesin or myosin. (adapted from R. D. Vale, Cell 112:467, 2003.)
Motors moving on cytoskeletal filaments can be classified into three types: myosins, kinesins and dyneins as shown in Figure 1. Though the diversity of these motors mirrors that of life more generally, we can attempt to classify them broadly into those that move on actin filaments and those that move on microtubules and according to the directionality of their motion. Both actin and microtubules have asymmetric filaments characterized by a plus-end and a minus-end. The motion of motors can in turn be characterized by the directions of their movement (i.e. plus-end or minus-end directed).
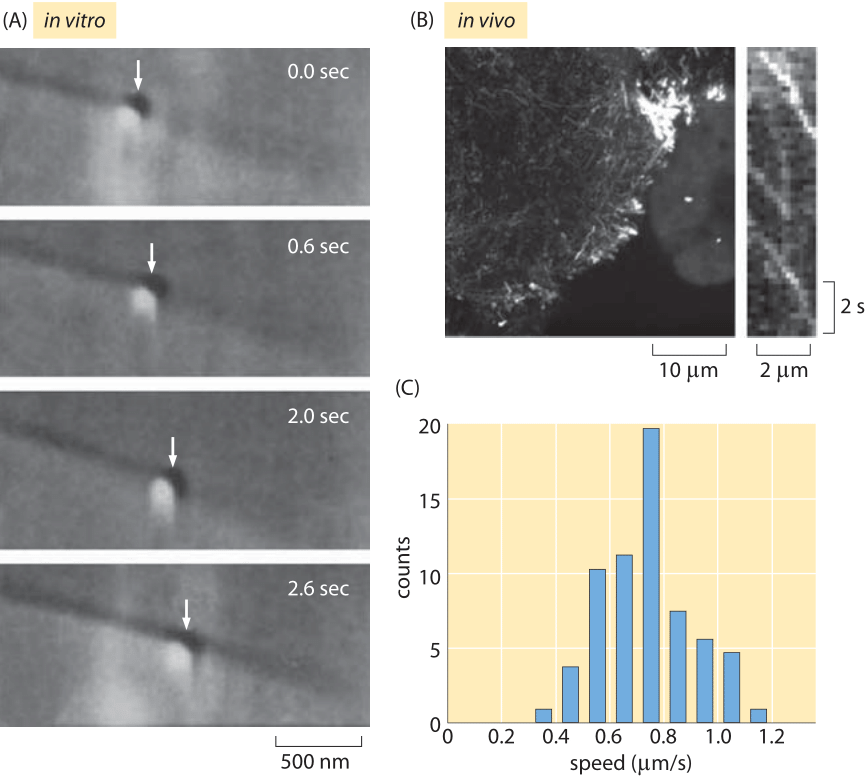
Fig. 2: Measuring kinesin motor velocities. (A) A glass bead coated with kinesin motors was brought in contact with a microtubule using an optical trap. Both the microtubule and the bead can be seen using DIC microscopy and the optical trap is visible as a slightly shiny spot around the bead. When the trap is shut off, the bead begins to move down the microtubule processively over several seconds. (B) Fluorescently labeled In vivo measurements of kinesin molecules fused to GFP. The kymograph shown on the right shows that the motors move roughly 2 microns in roughly 4 seconds. (C) Histogram of motor speeds from the measurements of ten cells like those made in (B). (A Adapted from S. M. Block et al., Nature 348:348, 1990, B, C Adapted from M. E. Tanenbaum et al., Cell 159:635, 2014.)
Once these motors have engaged their cargo, how fast can they move? For a single motor, as opposed to the collective motion of many motors that can engage a cargo simultaneously, there have been measurements made using single-molecule techniques in vitro as well as in vivo. Figure 2 shows examples of the kinds of microscopic observations that make these measurements possible. In now classic optical tweezers experiments, individual motors are tethered to a much larger bead that is then trapped using laser light. This trapping makes it possible to characterize the motor’s velocity as a function of the resistive force applied by the trapped bead. In many ways, the in vivo measurement of motor velocities is more conceptually straightforward since it involves essentially video microscopy of the motion of cargo within cells as shown in Figure 2B.
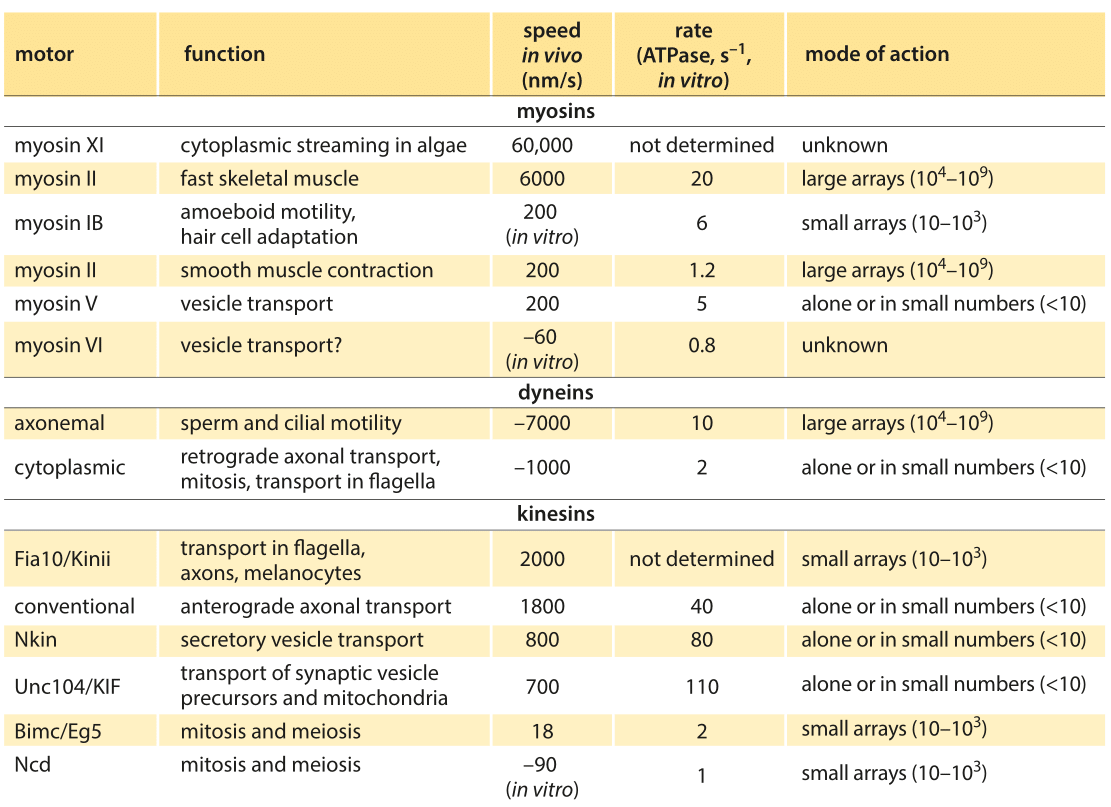
Table 1: Summary of experimental data on the dynamics of translational molecular motors. Based on BNID 106501 and 101506. Values were rounded to one significant digit. Negative speeds indicate movement towards the minus end of the filament.
Broadly speaking, translational motors move at rates somewhere between several tenths of a micron to several microns per second with some notable and very interesting outliers which broaden the distribution of motor speeds considerably. For example, conventional kinesins have an in vitro speed of 800 nm/s (BNID 101506) and an in vivo speed of 2000 nm/s. This directed motion is made up of individual steps of 8 nm length (BNID 101857), thus requiring about 100 steps per second to achieve such speeds in vitro, though clearly we are talking about the average response and the stochastic variations in these parameters are of great interest as well. After a characteristic duration of 100 steps the motor is released from the microtubule (BNID 103552). In every step one ATP molecule gets hydrolyzed releasing about 20 kBT of free energy. The force this can exert over the 8 nm step length is about 5 pN (assuming 50% efficiency, BNID 103008). A parametric spec sheet for these microscopic transport machines is given in Table 1.
Table 1: Summary of experimental data on the dynamics of translational molecular motors. Based on BNID 106501 and 101506. Values were rounded to one significant digit. Negative speeds indicate movement towards the minus end of the filament.
How large an object can be moved through a viscous environment at a 1 μm/sec velocity with this amount of force? Stokes’ law governs the relation of force (F) to velocity (V) in a fluid of viscosity η, through the relation F=6πηRV, where R is the radius of the object moving through the fluid. Plugging in the value for water, namely η=0.1 Pa s, we find a characteristic size of R=2 μm. This is about the upper limit on the size of an organelle, whereas most transport vesicles are significantly smaller than this bound. The value of the viscosity we used is that for water, in the highly crowded cellular interior the viscosity is higher but only by a factor of about 2-3 (BNID 105903, 103392).
Together, diffusion and motor-mediated active transport constitute two of the dominant mechanisms governing the lively comings and goings of molecules within cells. For active transport, evolution has resulted in a huge array of molecular motors with all sorts of elaborations that make it possible for them to move in different directions on different kinds of filaments while pulling along cargos which are themselves of a great diversity. Further, these motors are engaged in all sorts of dynamic activities within cells that do not relate to the transport of cargo at all, but rather, endow cells with their dynamism when separating chromosomes, moving around or separating in two.
The biophysical study of molecular motors helps clarify a seemingly magical sleight of hand, namely, how does the hydrolysis of phosphate bonds of diameter smaller than one nm get spatially amplified to entail a movement of your hand over a distance of order centimeters? The step of the myosin motor transforms the <1 nm phosphate bond severing to a movement two orders of magnitude longer of about 36 nm across a half period of the actin filament. This same action happening in a concerted direction in 104-105 sarcomeres per muscle amplifies the movement to the level of millimeters. Finally, the anatomy of the arm and its muscles gives the final leveraging to the domain of centimeters. With many biophysicists clarifying each of these steps in ever more rigorous detail, the micro to macro magic is demystified, as described in the book “Mechanisms of motor proteins and the cytoskeleton” by Jonathan Howard.

