How many ribosomal RNA gene copies are in the genome?
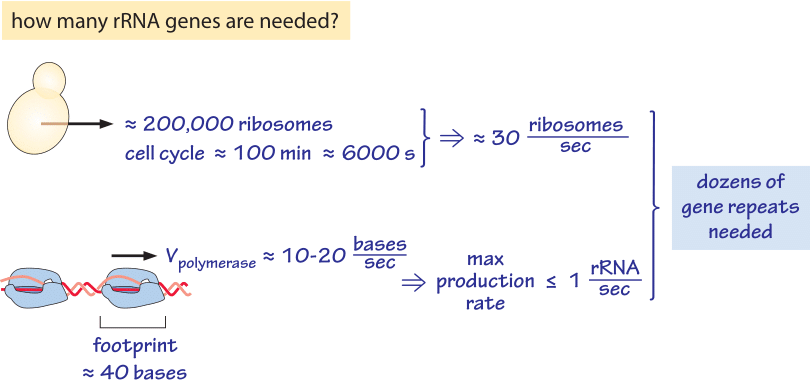
Figure 1: Back of the envelope estimation of the number of rRNA genes copies (repeats) needed in a budding yeast cell to supply the needed ribosome production rate under fast growth conditions. Knowledge of the rough number of ribosomes and the polymerase rate leads to several dozens of copies needed of the genes.
rRNA is the ribosomal RNA, a major constituent of the ribosome, accounting for about 2/3 of its mass (BNID 100119). In an earlier vignette on “How many ribosomes are in a cell?”, we discussed the large number of ribosomes required just to keep the steady pace of protein production moving. As a result of this high demand for protein production, under many growth conditions, one copy of the rRNA gene will not be enough to supply the ribosomal needs for cell growth even if that locus is being transcribed as fast as possible. Consider the budding yeast as depicted in Figure 1. Under fast exponential growth it is estimated to harbor about 200,000 ribosomes (BNID 100267). For a cell cycle time of 100 minutes, these cells need to produce ≈30 rRNA per second just to keep up with the demand for new ribosomes. Can the cell achieve this production rate with one gene copy? In yeast, transcription is performed by the RNA polymerase at an average speed of 10-20 bp/sec (BNID 103012, 103657). The polymerase size, or footprint over the DNA, is about 40bp (BNID 107873). Even if these genes were packed with RNA polymerase in a sequential array like cars in a traffic jam and all were moving at maximal speed, the net transcription rate would still be less than 1 rRNA/s. We thus need tens of copies of the rrna gene to enable enough transcription to supply new ribosomes at the necessary rate. Indeed, budding yeast is known to have about 150 rrn (encoding the rRNA) gene copies (BNID 101733, 100243). As an aside we note how these rrn genes are suggested to be important players in causing aging in budding yeast (Sinclair & Guarente, Cell 91:1033, 1997).
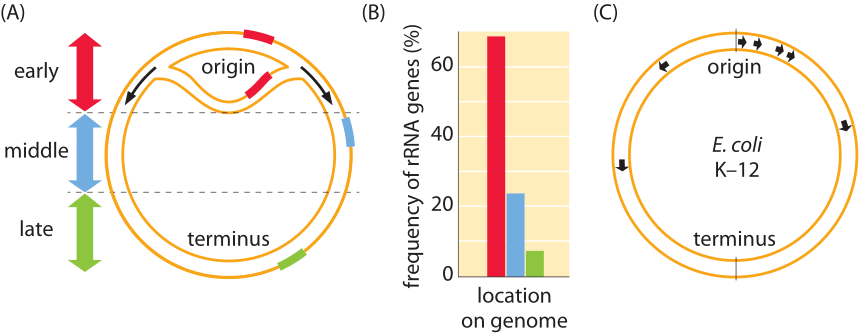
Figure 2: Ribosomal RNA genes across microbial genomes. (A). Frequency in each third of the chromosome of rDNA operons in 68 bacterial genomes. (B). Locations of the rRNA genes on the circular genome of E. coli. Note that all copies of the RNA genes are on the leading strand and none are on the lagging strand. (A, adapted from E. P. C. Rocha Microbiology 150:1609, 2004. B, Adapted from M Nomura PNAS;96:1820, 1999).
Even in E. coli where very few genes appear with more than one copy per chromosome there are 7 copies of the rRNA genes as shown in Figure 2 (BNID 102219). A qualitative way to think about why there must be several copies of the rRNA gene is that rRNA is not translated so there is no second amplification step in translation as there would be for many proteins. Therefore, the only way to achieve the necessary concentration of ribosomes is to have many gene copies. The number of ribosomes in cells has been suggested to be limited by the number of operon copies with ribosomal proteins being regulated to match the synthesis rate of rRNA. The importance of the number of copies of rrn genes has been tested in a study in E. coli where rrn operons were deleted and the resulting growth rate was measured as a function of the rrn copy number for a range of 1-7 copies. With less than 6 copies there was a significant decrease in growth rate. In other experiments, extra copies beyond 7 have also been shown to be detrimental to growth, possibly because of an increase in the diffusion times as the cytoplasm becomes ever more packed with ribosomes (T. Asai et al, J Bact. 181:3803, 1999; D. A. Schneider & R. L. Gourse, J Bact. 185:6185, 2003; A. D. Tadmor & T. Tlusty, PLOS comp bio, 4:5, 2008).
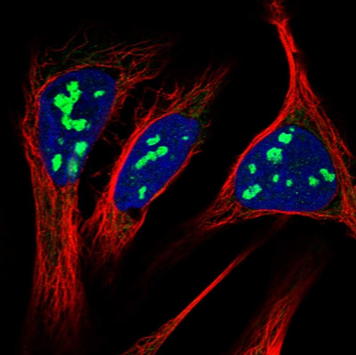
Figure 3: Visualizing the nucleoli, the location of ribosomal RNA transcription and assembly in the human cell line U-2 OS. Immunofluorescent staining for the gene RRP15 that is located in the nucleoli and functions in the maturation of the ribosomal subunit. Staining of gene (nucleoli) in green, with additional staining of nucleus in blue and microtubules in red. From: http://www.proteinatlas.org/
ENSG00000067533/subcellular
The number of rRNA copies in E. coli has apparently remained constant at 7 copies per genome over the ≈150 million years since it diverged from S. typhimurium (BNID 107087, 107867). As E. coli can have nested replication forks resulting in multiple DNA replication operons close to the origin, under fast growth rates there will be more copies of the origin. Indeed, the rrn operons tend to be close to the origin and the number of copies per cell can be much higher than 7 – at high growth rate this number can be as high as 36 (BNID 102359). Advances in genomics allow us to retrieve the number of rRNA genes of hundreds of organisms and these numbers are now summarized in databases (BNID 104390). The databases tables show how the number of copies tends to be higher in faster growing cells. Information on their location along the genome can show for example how common is the tendency to cluster near the origin as shown in Figure 2B. One can also observe the tendency of rRNA to be coded on the leading strand (arrow direction facing away from the origin in Figure 1A) rather than on the lagging strand in replication as further discussed in the caption. This can be understood through the “collision avoidance” model. When the replication fork overtakes a transcribing RNA polymerase moving in the same direction there is replication slow down until transcription ends. When the RNA polymerase is moving in the direction opposite to the fork advance there is a head on collision that leads to a much more problematic replication arrest and transcription abortion. Avoidance of these latter cases selects for locating RNA genes on the leading strand (E. P. C. Rocha, Ann. Rev. Genet. 42:211, 2004).
How does this question of number of ribosomal RNA genes play out in the case of Homo sapiens? Humans carry about 200 copies of the rrn genes in their genome (BNID 107865), these are organized in 5 clusters known as the nucleolus organizers each containing multiple copies of the rRNA operon. In phase microscopy of human cells these areas of extensive transcriptional activity known as nucleoli are vividly seen as black dots inside the nucleus and even more strikingly in fluorescent microscopy as seen in Figure 3. Clearly, in organisms ranging from bacteria to humans, the number of ribosomes is a critical cellular parameter and one of the main ways that it is regulated is through the gene copy number itself.

