What are the concentrations of cytoskeletal molecules?
Just as there is a battery of macromolecules that participate in the flow of information between proteins and DNA, there is also a wide collection of different molecules that dictate when and where the molecules of the cytoskeleton will be assembled into the filamentous networks that crisscross cells. When thinking about the question of cell motility, leading this cast of molecular players is the protein actin, a soluble protein with a run-of-the-mill ≈40 kDa mass but which forms rigid filamentous assemblies with long persistence lengths of about 10 µm (BNID 106830) that are crucial for propelling cells forward.
As shown in the vignette on cytoskeletal sizes, the leading edge of a motile cell such as a keratocyte is characterized by a dense and branched network of actin filaments which create protrusions such as filopodia and lamellipodia. These protrusions are peppered with sites of adhesion between the cell and external solid substrate. These sites of adhesion have a characteristic diameter of 100-300 nm (BNID 102267) and an average lifetime of 20 s (BNID 102266)), serving as anchors for the mesmerizing cellular dynamics revealed in time-lapse images of motile cells crawling on surfaces.
How much actin does it take to set up such a network? Similarly, how many attendant proteins are there to make sure that such filaments are “constructed’’ at the right time and place? One way to begin to answer such questions is through simple estimates based upon inspecting electron microscopy images of typical filaments at the leading edge of motile cells. Since the size of a typical monomer is roughly 5 nm and the filaments themselves are characterized by micron-scale lengths, each filament is made up of hundreds of actin monomers. Though electron microscopy images provide a compelling structural vision of the leading edge of a motile cell, they leave us wondering about the host of other molecular partners that control the spatiotemporal patterns of filament formation. Other methods (and cell types) have been used to take the molecular roll call of the many proteins implicated in cytoskeletal network formation.
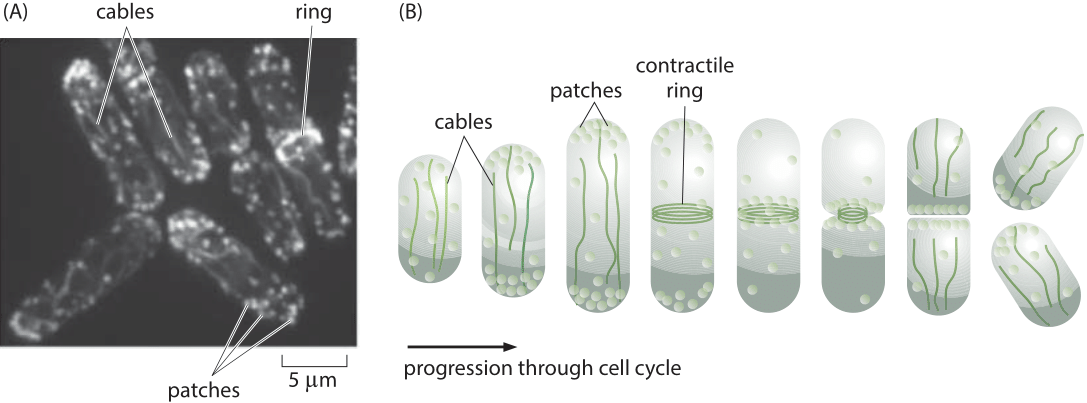
Figure 1: The actin cytoskeleton in fission yeast. (A) Fluorescence microscopy image of the various actin structures found in the fission yeast. (B) Schematic of the time variation of the distribution of actin over the cell cycle. During the cell division process, actin normally invested in patches and cables is retasked to forming the contractile ring. (Adapted from D. R. Kovar et al., Trends Cell Biol., 21:177, 2011.)
A powerful model system for investigating questions about the dynamics of the actin cytoskeleton is provided by the fission yeast Schizosaccharomyces pombe. One of the reasons that these eukaryotic cells are so useful is that their uses of actin are centered on the formation of three specialized classes of structures as shown in Figure 1. The first class of actin structure is that associated with intracellular transport, a signature feature of eukaryotes, and in fission yeast, it is the cargo-carrying molecular motors that move along this network of actin filaments that mediates this process. A second of the primary functions of the actin cytoskeleton is to mediate the fission process whereby one mother cell divides into two daughters through the formation of a contractile ring at the cell middle. Finally, actin is a key player in the endocytosis process where the formation of dense actin patches provides part of the force-generating machinery that makes membrane invaginations possible. These fission yeast cells were used to take a careful census of the actin cytoskeleton that gives a sense of the absolute numbers and concentrations of both actin and its accessory proteins.
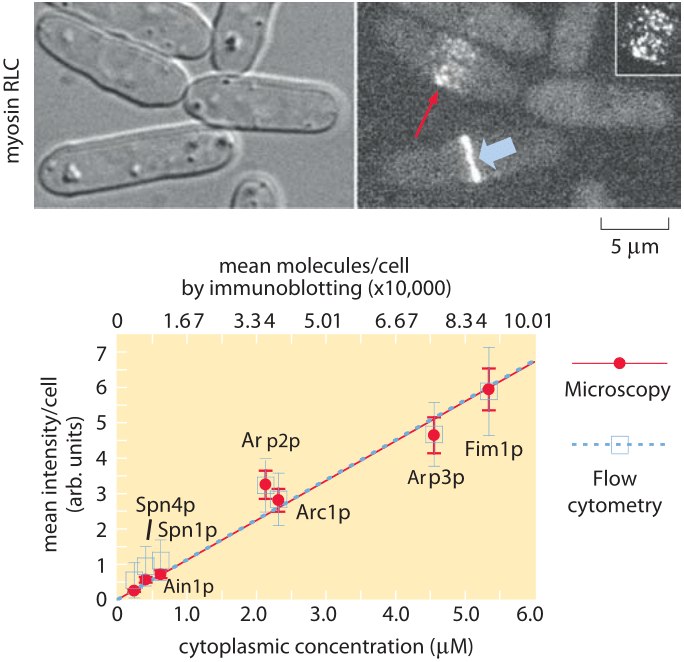
Figure 2: Molecular census of the actin cytoskeleton in fission yeast. (A) Phase contrast images of fission yeast cells. (B) Fluorescence images of myosin. (C) Calibration of the census. The number of molecules per cell as determined from immunoblotting shows a linear relation with the average fluorescence per cell. (Adapted from J.-Q. Wu and T. D. Pollard, Science, 310:310, 2005.)
To get a sense of the number of molecular copies of the cytoskeletal proteins and their various accessory proteins, systematic fusion of fluorescent proteins to each and every actin-related protein and calibration of the fluorescence signal using antibody techniques permitted a direct measurement of protein copy numbers as shown in Figure 2. Specifically, by measuring overall fluorescence levels and then exploiting calibration factors to convert intensities into molecular counts it was possible to determine the molecular census for an entire suite of actin-related proteins. As reported in Table 1, the numbers per cell range from just over 1 million copies of actin monomers per cell (about 1% of the proteome, making it one of the most abundant in the cell, see also the vignette on “What are the most abundant proteins in a cell?”) to somewhat less than 1000 copies of the actin filament capping protein formin.
What are the host of different actin-related proteins all for? One of the hallmark features of “living matter’’ is the exquisite control that is exercised over cellular processes. That is, most biological processes only happen when and where they are supposed to. In the case of actin polymerization, what this means is that there is a battery of control proteins for coordinating the actin polymerization process. For example, proteins that cap monomers thus forbidding them from participating in filament formation, proteins that communicate with membrane lipids that tell actin to form filaments near these membranes in order to form the protrusions at the leading edge, proteins that bind to preexisting filaments and serve as branching sites to send off new filaments in a different direction, etc. As seen in the table, there are more than 50 such proteins and they occur with different concentrations covering a range of about 100 fold from tens of nM to several µM.
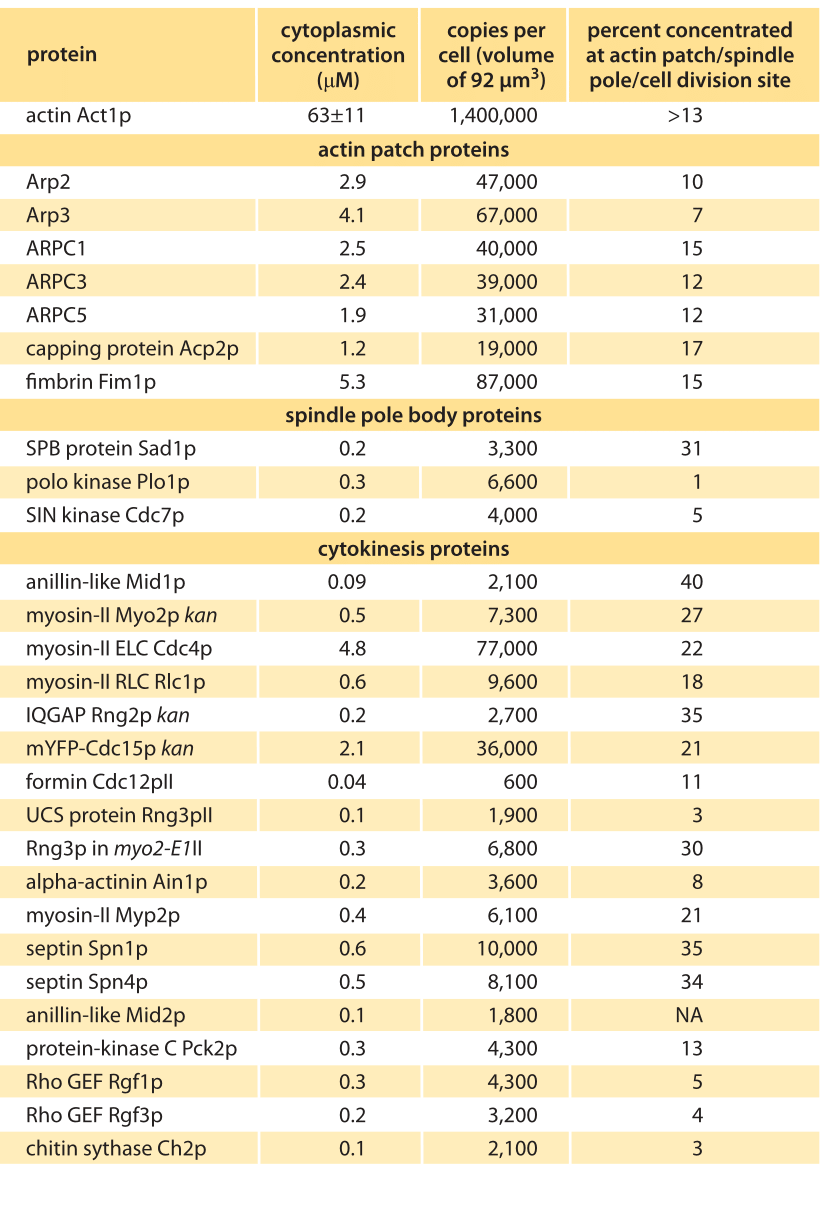
Table 1 – Concentrations of actin and actin related proteins in S. pombe from Wu & Pollard, Science, 310:310, 2005. Values are rounded to two significant digits.
How can we rationalize the numbers as detailed in the Table? One of the immediate impressions that comes from inspection of the data is that there are in some cases orders of magnitude differences in the quantities of different proteins. For example, while there are in excess of a million actin monomers, there are only roughly 50,000 copies of the protein complex that regulates the actin cytoskeleton, Arp2/3, and only 600 copies of the regulatory protein formin. Of course, these numbers make intuitive sense since a given filament might only be decorated by one Arp2/3 complex or formin dimer. These abundances might be further reasoned out by imagining several different categories of molecules. First, it is not surprising that actin is in a category by itself since it is the fundamental building block for constructing the long filaments involving tens to thousands of monomers each. The second category of molecules are those that are required at a stoichiometry of one or a few per filament or patch, such as the capping and branching proteins. These would be expected to be found with tens of thousands of molecules per cell as we discuss below. Finally, we might expect that regulatory proteins could be found in quantities of less than one copy per filament. Referring to Table 1 we see that most factors such as motor components (myosin) or branching (Arp) come with copy numbers in the many thousands while regulatory proteins (kinases) are in the few thousands. To think up the number of filaments and monomers it is useful to think of the interphase stage of the cell cycle when much of the actin is tied up in the formation of several hundred actin patches distributed across the cell, with each such patch containing more than 100 small filaments built up from 10-100 monomers. To construct all of these patches requires more than 500,000 actin monomers, corresponding to nearly half of the pool of utilized monomers. During mitosis, this balance is shifted since at this stage in the cell cycle, nearly half of the actin is now invested in constructing the contractile ring at the center of the cell. This ring is responsible for pinching the two daughter cells apart. The actin invested in the construction of this ring can be reasoned out by noting that there are roughly 2000 filaments making up these rings, with each such ring roughly ½ µm in length, implying that hundreds of thousands of monomers are implicated in the formation of these rings.
These are only several examples of the rich and complex cytoskeletal architectures found in living cells. As can be seen during cell division for eukaryotic cells, there is also an equally fascinating network of microtubule filaments that are key to separating the newly formed chromosome copies into the daughter cells. Microtubules also form molecular highways on which traffic is shuttled around by cargo-carrying molecular motors. Similar rationale might be provided for the microtubule-related census, though current experimental attempts to characterize the microtubule cytoskeleton lag behind efforts on the actin-based system.
All told, the cytoskeleton is one of the most critical features of cellular life and just as we need to know about the concentration of transcription factors to understand how they regulate genetic decision making, the concentrations of cytoskeletal proteins and their accessory factors is critical to developing a sense of the highly orchestrated dynamics found in cells.

