What is the error rate in transcription and translation?
The central dogma recognizes the flow of genomic information from the DNA into functional proteins via the act of transcription, which results in synthesis of messenger RNA, and the subsequent process of translation of that RNA into the string of amino acids that make up a protein. This chain of events is presented in textbooks as a steady and deterministic process, but is, in fact, full of glitches in the form of errors in both the incorporation of nucleotides into RNA and amino acid incorporation in proteins. In this vignette we ask: how common are these mistakes?
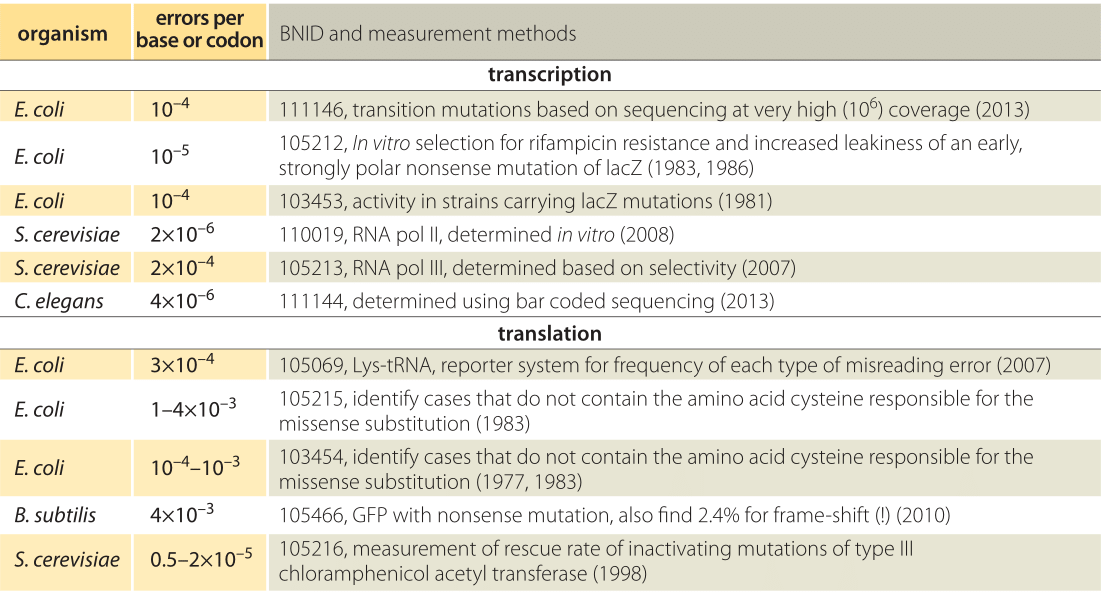
Table 1: Error rates in transcription and translation. For transcription the error rates are given per base whereas for translation the error rates are per codon, i.e. amino acid.
One approach to measuring the error rate in transcription is to use an E. coli mutant carrying a nonsense mutation in lacZ (i.e. one that puts a premature stop codon conferring a loss of function) and then assay for activity of this protein which enables utilization of the sugar lactose. The idea of the experiment is that functional LacZ will be produced through rare cases of erroneous transcription resulting from a misincorporation event that bypasses the mutation. The sensitivity of the assay makes it possible to measure this residual activity due to “incorrect” transcripts giving an indication of an error rate in transcription of ≈10-4 per base (BNID 103453, Table 1), which in this well orchestrated experiment changed the spurious stop codon to a codon responsible instead for some other amino acid, thus resurrecting the functional protein. Later measurements suggested a value an order of magnitude better of 10-5 (BNID 105212 and Ninio, Biochimie, 73:1517, 1991). Ninio’s analysis of these error rates led to the hypothesis of an error correction mechanism termed kinetic proofreading, paralleling a similar analysis performed by John Hopfield for protein synthesis. Recently, GFP was incorporated into the genome in the wrong reading frame enabling the study of error rates for those processes resulting in frame-shifts in the bacterium B. Subtilis. A high error rate of about 2% was observed (BNID 105465) which could arise at either the transcriptional or translational levels as both could bypass the inserted mutation. The combined error rate for the frame-shift is much higher than estimated values for substitution mutations indicating that the prevalence and implications of errors are still far from completely understood. Like with many of the measurements described throughout our book, often, the extremely clever initial measurements of key parameters have been superseded in recent years by the advent of sequencing-based methods. The study of transcription error rates is no exception with recent RNA-Seq experiments making it possible to simply read out the transcriptional errors directly, though these measurements are fraught with challenges since sequencing error rates are comparable to the transcriptional error rates (10-4-10-5) that are being measured.
The error rate of RNA polymerase III, the enzyme that carries out transcription of tRNA in yeast has also been measured. The authors were able to tease apart the contribution to transcriptional fidelity arising from several different steps in the process. First, there is the initial selectivity itself. This is followed by a second error-correcting step that involves proofreading. The total error rate was estimated to be 10-7 which should be viewed as a product of two error rates, ≈10-4 arising from initial selectivity and an extra factor of ≈10-3 arising from proofreading (105213, 105214). Perhaps the best way to develop intuition for these error rates is through an analogy. An error rate of 10-4 corresponds to the authors of this book making one typo every several vignettes. An error rate of 10-7 corresponds more impressively to one error in a thousand-page textbook (almost an impossibility for most book authors….).
Error rates in translation (10-4-10-3) are generally thought to be about an order of magnitude higher than those in transcription (10-5-10-4) as roughly observed in Table 1. For a characteristic 1000 bp/300 aa gene this suggests on the order of one error per 30 transcripts synthesized and one error per 10 proteins formed. Like with measurements of errors in transcription, one of the ways that researchers have gone about determining translational error rates is by looking for the incorporation of amino acids that are known to not be present in the wild-type protein. For example, a number of proteins are known not to have any cysteine residues. The experiment then consists in using radioactive isotopes of sulfur present in cysteine and measuring the resulting radioactivity of the newly synthesized proteins, with rates in E. coli using this methodology yielding mistranslation rates of 1-4 x 10-3 per residue. One interpretation of the evolutionary underpinnings of the lower error rate in transcription than in translation is that an error in transcription would lead to many erroneous protein copies whereas an error in translation affects only one protein copy. Moreover, the correspondence of 3 nucleotides to one amino acid means that mRNA messages require higher fidelity per “letter” to achieve the same overall error rate. Note also that in addition to the mistranslation of mRNAs, the protein synthesis process can also be contaminated by the incorrect charging of the tRNAs themselves, though the incorporation of the wrong amino acid on a given tRNA has been measured to occur with error rates of 10-6.
A standing challenge is to elucidate what limits the possibility to decrease the error rates in these crucial processes in the central dogma even further, say to values similar to those achieved by DNA polymerase. Is there a biophysical tradeoff in play or maybe the observed error rates have some selective advantages?

