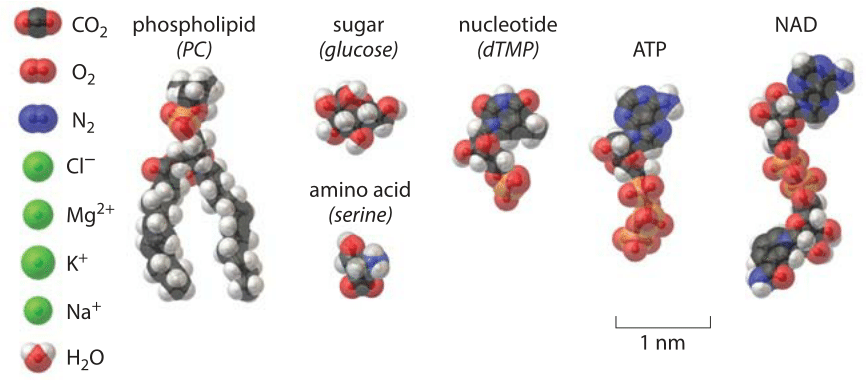
How big are biochemical nuts and bolts?
The textbook picture of the molecules of life is dominated by nucleic acids and proteins, in no small measure because of their fascinating linkage through the processes of the central dogma. On the other hand, this picture is terribly distorted biochemically because many of the key reactions even in the central dogma would not happen at all were it not for a host of biochemical allies such as water and the many ions that are needed as cofactors for the enzymes that make these reactions go. Further, we cannot forget the substrates themselves, namely, the nucleotides and amino acids from which the famed nucleic acids and proteins are constructed. Energizing all of this busy activity are small sugar molecules, energy carriers such as ATP and other metabolites. In this vignette, we take stock of the sizes of the many biochemical “nuts and bolts” that provide the molecular backdrop for the lives of cells as shown in Figure 1.
Probably the single most important biochemical nut and bolt of them all is water. It is no accident that the search for life beyond Earth often begins with the question: is there water? Though part of the reason for this might be a lack of imagination about what other life-supporting chemistries might look like, the simplest reason for this obsession with water is that without it, life as we know it could not exist. One of the easiest ways for us to characterize the size of a water molecule which is a convenient standard molecular ruler for biology is by reference to the roughly 0.1 nm bonds (BNID 106548) between its hydrogen and oxygen atoms. Since water molecules are not spherically symmetric it is hard to assign an effective radius to such a molecule. As another estimate for the size of a water molecule we appeal to the mean spacing between such molecules by using the density of water. In particular, given that there are 55 moles of water per liter, we find the volume of a water molecule to be 0.03 nm3, and the mean spacing between molecules to be roughly 0.3 nm (BNID 106548). We will also find it convenient to use the 18 Da mass of water as a way of comparing the sizes of these various molecular players.
We all come from the ocean. Despite our human dependence on fresh water for drinking and maintaining the many plants and animals that feed us, real biological water bears the signature of our watery origins in the ocean. Our first impression on hopping into the ocean (besides that it is cold!) is likely the salty taste it leaves on our tongues. A simple estimate of the saltiness of the ocean can be garnered from remembering that a kilogram of water has roughly 55 moles of water molecules (i.e. 1000 g/18 g/mole). This same seawater has roughly 1 mole of salt (BNID 100802) meaning that 1 out of every 55 molecules is an ion. If we look within cells, we find a number of different ions such as H+, Na+, K+, Mg2+ and Cl– that add up to about a quarter of the concentration of sea water as discussed in the vignette “What are the concentrations of different ions in cells?”. The sizes of these ions can be captured by the so-called ionic radii which are given by Na+ = 0.09 nm, K+ = 0.13 nm, Mg2+=0.07 nm and Cl– = 0.18 nm (BNID 108517, 104162, 109742, 109743, 103950). These ionic radii reveal the so-called “bare” ionic radius whereas the hydrated ionic radius is usually much larger, and more similar among ions, at 0.3-0.4 nm (BNID 108517). These surrounding water molecules are exchanged on the micro to nanosecond time timescale (BNID 108517). The hydrated ions radii are shown to scale next to other nuts and bolts of the cell in Figure 1.
To build up the nucleic acids and proteins of the cell requires molecular building blocks. The nucleotides that are the building blocks of nucleic acids have a mass of ≈300 Da. Their physical size is compared to water in the gallery shown in Figure 1, though we can also get a feel for this size by remembering that the DNA double helix has a radius of roughly 1 nm and an average spacing between bases along the chain of 1/3 nm. This means that a plasmid of say 10 kbp will have a circumference of about 3000 nm, i.e. a diameter of about 1 micron. The common depiction of plasmids as small circles inside a bacteria are easy to understand but do not do justice to the physical size of plasmids. Indeed plasmids in cells must be curled up to fit in. The amino acids that make up proteins range in size from the tiny glycine with a molecular mass of roughly 75 Da to the 204 Da mass of tryptophan, the largest of the naturally occurring amino acids. Their respective lengths vary from 0.4 to 1 nm (BNID 106983). Adopting a mass of 100 Da per aa in a protein polymer serves as a very useful and calculationally convenient rule of thumb. Here too, the sizes of the amino acids with respect to a water molecule are shown in Figure 1.
All of this emphasis on nucleic acids and proteins can lead us to forget the critical role played in the lives of cells both by lipids and sugars. The emerging field of lipidomics has shown that just as there is immense diversity in the character of the many proteins that inhabit cells, the membranes of the cells and their organelles are similarly characterized by widely different concentrations of an entire spectrum of different lipids (see the vignette “What lipids are most abundant in membranes?”). In simplest terms, the lipids making up these membranes have a cross sectional area of between 0.25 and 0.5 nm2, and a length of order 2 nm as shown in Figure 1. More generally, the lengths of the lipid chains are dictated by the number of carbons they contain with a rule of thumb that L=a+b x n, where n is the number of carbons in the tail and a and b are constants depicting, respectively, the terminal group size outside the carbon chain and the length extension per carbon atom. The masses of lipids are between 700 and 1000 Da as a general rule.
Cellular life is powered by a number of other key molecules besides those discussed so far. To grow new cells, biologists use various kinds of growth media, but some of the most standard ingredients in such media are sugars such as glucose. With a chemical formula of C6H12O6, glucose has a molecular mass of 180 Da. Structurally, the glucose molecule is a 6-membered ring as shown in Figure 1 with typical carbon-carbon bond lengths of ≈0.15 nm and an overall molecular size of roughly 1 nm as measured by the long axis of the cyclic form or the length of the open chain form (BNID 110368, 106979). Once sugars are present within a cell, they can be remodeled to build the carbon backbones of molecules such as the nucleotides and amino acids described above, and also for the synthesis of key energy carriers such as ATP. The size of ATP (effective diffusion diameter of ≈1.4 nm, BNID 106978) is compared to the rest of the biochemical nuts and bolts in Figure 1. ATP, is a nucleotide adapted to piggyback energized phosphate groups, and has a molecular mass of roughly 500 Da. The other major energy sources are electron donors with NADP being the prime shaker and mover with a mass of 744 Da and a length of about 2.5 nm (BNID 106981).
In summary, if one has to carry one round number to utilize for thinking about sizes of small building blocks such as amino acids, nucleotides, energy carriers etc., 1 nm is an excellent rule of thumb.