How big are genomes?
Genomes are now being sequenced at such a rapid rate that it is becoming routine. As a result, there is a growing interest in trying to understand the meaning of the information that is stored and encoded in these genomes and to understand their differences and what these differences say about the evolution of life on Earth. Further, it is now even becoming possible to compare genomes between different individuals of the same species, which serves as a starting point for understanding the genetic contributions to their observed phenotypes. For example, in humans, so called genome-wide association studies associate variations in genetic makeup with susceptibility to diseases such as diabetes and cancer.
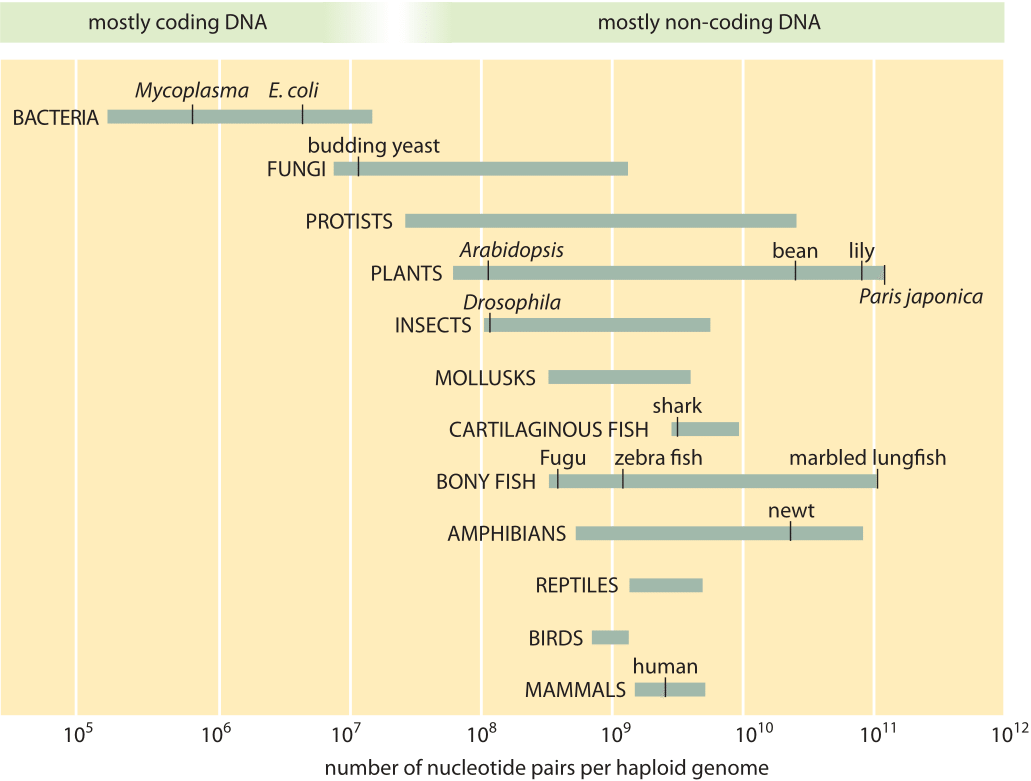
Naively, the first question one might ask in trying to take stock of the information content of genomes would be how large are they? Early thinking held that the genome size should be directly related to the number of genes it contains across the whole tree of life. This was strikingly refuted by the similarity in the number of protein coding genes in genomes of very different sizes, one of the unexpected results of sequencing many different genomes from organisms far and wide. For example, as shown in Table 1, Caenorhabditis elegans (a nematode) has a very similar number of protein coding genes to that of human or mouse (≈ 20,000) even though their genomes vary in size by over 20 fold. As shown in Figure 1, the range of genome sizes runs from the 0.16 Mbp for the endosymbiotic Candidatus ruddii to the ≈150 Gbp (BNID 110278) for the enormous genome of the plant Paris Japonica, revealing a million fold difference in genome size. An often-cited claim for a world record genome size at 670 Gbp for the amoeba Polychaos dubium is considered dubious as it used 1960s methods that analyzed the whole cell rather than single nuclei. Because of this approach, the result could be muddled by including contributions from mitochondrial DNA, possible multiple nuclei and anything the amoeba recently engulfed (BNID 104470). At the other extreme of small genome sizes, viral genomes are in a class of their own where sizes are usually considerably smaller than the smallest bacterial genome with many of the most feared RNA viruses having genomes that are less than 10 kb in length.
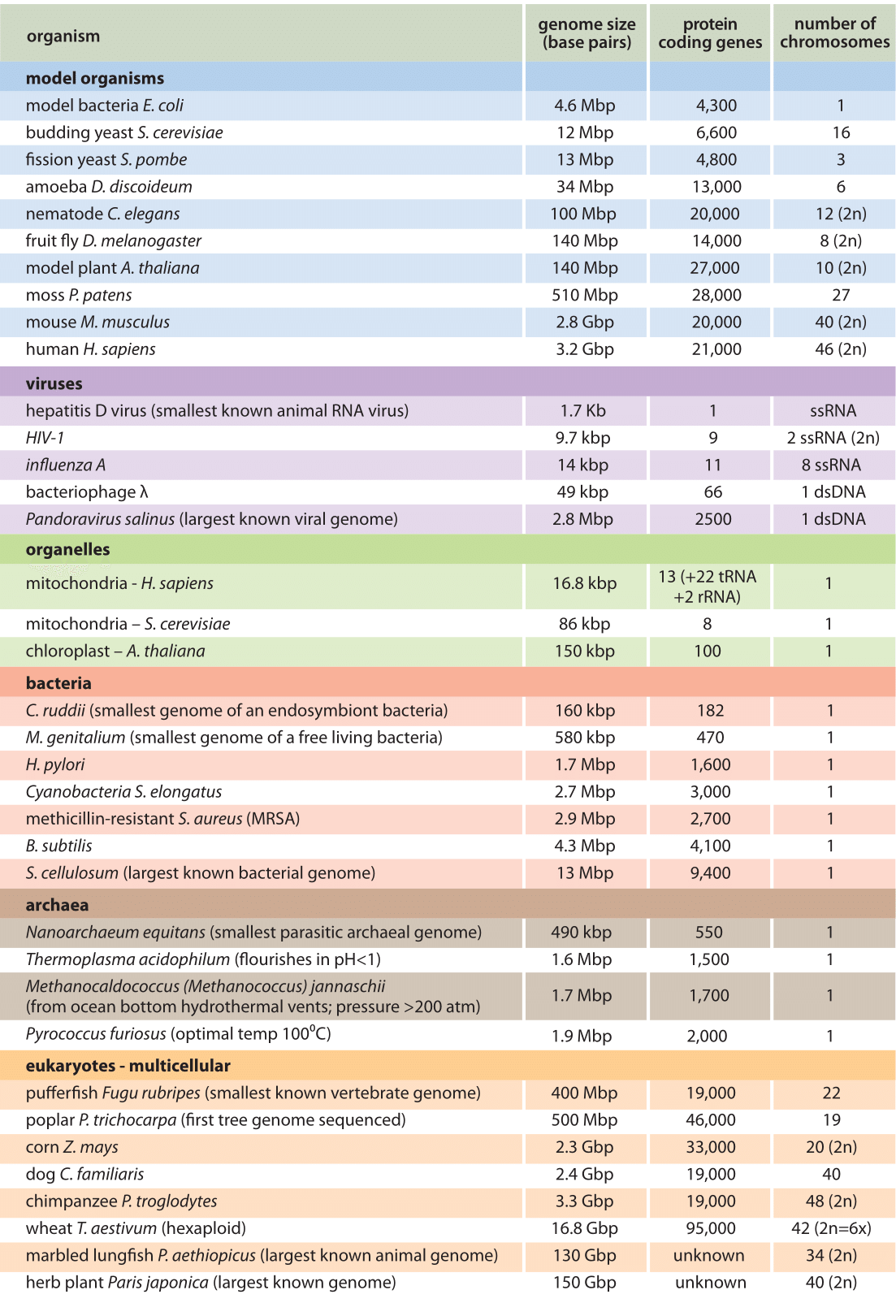
Table 1 – Genomic census for a variety of selected organisms. The table features the genome size, current best estimate for number of protein coding genes and number of chromosomes. Genomes often also include extra-chromosomal elements such as plasmids that might not be indicated in the genome size and number of chromosomes. The number of genes is constantly under revision. The numbers given here reflect the number of protein coding genes. tRNA and non coding RNAs, many of them still to be discovered, are not accounted for. Bacterial strains often show significant variations in genome size and number of genes among strains. Values were round to two significant digits.
What is the physical size of these DNA molecules? Converting the length as measured in base pair units to physical length of the fully stretched out DNA molecule can be carried out by noting that the distance between bases along the DNA strand is ≈0.3 nm (BNID 100667). For the human genome with its length of ≈3Gbp, this conversion tells us that each of our more than 1013 cells harbors roughly a meter of DNA. Remarkably, each cell in our body has to compress this one meter’s worth of DNA into a nuclear volume with a radius of only a few microns. There is actually double trouble as our cells are diploid meaning that each nucleus has to pack roughly 2 full meters worth of DNA. To carry out this extreme compaction requires architectural proteins such as histones and much dexterity in reading the stored information during transcription. Similarly in bacteria, every operon such as the Lac operon, if it was stretched in a straight line, would by itself traverse the whole length of the bacterium.
Figure 1 and Table 1 give examples of different genome sizes with the ambition of illustrating some of the useful and well known model organisms, some of the key outliers characterized by genomes that are either extraordinarily small or large and examples which are particularly exotic. For some of the largest genomes, such as the record holder of the animal kingdom, the marbled lungfish, sequencing is not yet available. Older methods of measuring DNA in bulk refer to the genome size through the C-value, representing the amount of DNA and thus genome length without regard to its specific sequence. The next vignettes now take up the question of how many chromosomes and genes are present in these various genomes and whether there are any useful rules of thumb for predicting the gene number on the basis of genome size.