What is the protein to mRNA ratio?
The central dogma hinges on the existence and properties of an army of mRNA molecules that are transiently brought into existence in the process of transcription and often, shortly thereafter, degraded away. During the short time that they are found in a cell, these mRNAs serve as a template for the creation of a new generation of proteins. The question posed in this vignette is this: On average, what is the ratio of translated message to the message itself?
Though there are many factors that control the protein-mRNA ratio, the simplest model points to an estimate in terms of just a few key rates. To see that, we need to write a simple “rate equation” that tells us how the protein content will change in a very small increment of time. More precisely, we seek the functional dependence between the number of protein copies of a gene (p) and the number of mRNA molecules (m) that engender it. The rate of formation of p is equal to the rate of translation times the number of messages, m, since each mRNA molecule can itself be thought of as a protein source. However, at the same time new proteins are being synthesized, protein degradation is steadily taking proteins out of circulation. Further, the number of proteins being degraded is equal to the rate of degradation times the total number of proteins. These cumbersome words can be much more elegantly encapsulated in an equation which tells us how in a small instant of time the number of proteins changes, namely,
p(t+Δt)=p(t)-αp(t)Δt+βmΔt
where α is the degradation rate and β is the translation rate (though the literature is unfortunately torn between those who define the notation in this manner and those who use the letters with exactly the opposite meaning).
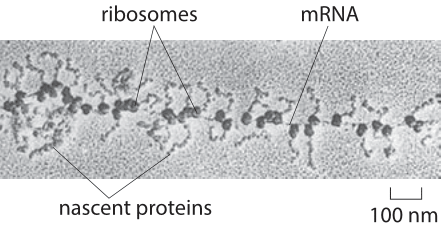
Figure 1: Ribosomes on mRNA as beads on a string (from: http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html)
We are interested in the steady state solution, that is, what happens after a sufficiently long time has passed and the system is no longer changing. In that case dp/dt=0=βm-αp. This tells us in turn that the protein to mRNA ratio is given by p/m = β/α. We note that this is not the same as the number of proteins produced from each mRNA, this value requires us to also know the mRNA turnover rate which we take up at the end of the vignette. What is the value of β? A rapidly translated mRNA will have ribosomes decorating it like beads on a string as captured in the classic electron micrograph shown in Figure 1. Their distance from one another along the mRNA is at least the size of the physical footprint of a ribosome (≈20 nm, BNID 102320, 105000) which is the length of about 60 base pairs (length of nucleotide ≈0.3 nm, BNID 103777), equivalent to ≈20 aa. The rate of translation is about 20 aa/sec. It thus takes at least one second for a ribosome to move along its own physical size footprint over the mRNA implying a maximal overall translation rate of β=1 s-1 per transcript.
The effective degradation rate arises not only from degradation of proteins but also from a dilution effect as the cell grows. Indeed, of the two effects, often the cell division dilution effect is dominant and hence the overall effective degradation time, which takes into account the dilution, is about the time interval of a cell cycle, τ. We thus have α = 1/τ.
In light of these numbers, the ratio p/m is therefore 1 s-1/(1/τ)= τ. For E. coli, τ is roughly 1000 s and thus p/m~1000. Of course if mRNA are not transcribed at the maximal rate the ratio will be smaller. Let’s perform a sanity check on this result. Under exponential growth at medium growth rate E. coli is known to contain about 3 million proteins and 3000 mRNA (BNID 100088, 100064). These constants imply that the protein to mRNA ratio is ≈1000, precisely in line with the estimate given above. We can perform a second sanity check based on information from previous vignettes. In the vignette on “What is heavier an mRNA or the protein it codes for?” we derived a mass ratio of about 10:1 for mRNA to the proteins they code for. In the vignette on “What is the macromolecular composition of the cell?” we mentioned that protein is about 50% of the dry mass in E. coli cells while mRNA are only about 5% of the total RNA in the cell which is itself roughly 20% of the dry mass. This implies that mRNA is thus about 1% of the overall dry mass. So the ratio of mRNA to protein should be about 50 times 10, or 500 to 1. From our point of view, all of these sanity checks hold together very nicely.

Figure 2: Simultaneous measurement of mRNA and protein in E. coli. (A) Microscopy images of mRNA level in E. coli cells. (B) Microscopy images of protein in E. coli cells. (C) Protein copy number vs mRNA levels as obtained using both microscopy methods like those shown in part (A) and using sequencing based methods. From Taniguchi et al. Science. 329, 533 (2010).
Experimentally, how are these numbers on protein to mRNA ratios determined? One elegant method is to use fluorescence microscopy to simultaneously observe mRNAs using fluorescence in-situ hybridization (FISH) and their protein products which have been fused to a fluorescent protein. Figure 2 shows microscopy images of both the mRNA and the corresponding translated fusion protein for one particular gene in E. coli. Figure 2C shows results using these methods for multiple genes and confirms a 100- to 1000-fold excess of protein copy numbers over their corresponding mRNAs. As seen in that figure, not only is direct visualization by microscopy useful, but sequence-based methods have been invoked as well.
For slower growing organisms such as yeast or mammalian cells we expect a larger ratio with the caveat that our assumptions about maximal translation rate are becoming ever more tenuous and with that our confidence in the estimate. For yeast under medium to fast growth rates, the number of mRNA was reported to be in the range of 10,000-60,000 per cell (BNID 104312, 102988, 103023, 106226, 106763). As yeast cells are ≈50 times larger in volume than E. coli, the number of proteins can be estimated as larger by that proportion, or 200 million. The ratio p/m is then ≈2×108/2×104≈104, in line with experimental value of about 5,000 (BNID 104185, 104745). For yeast dividing every 100 minutes this is on the order of the number of seconds in its generation time, in agreement with our crude estimate above.
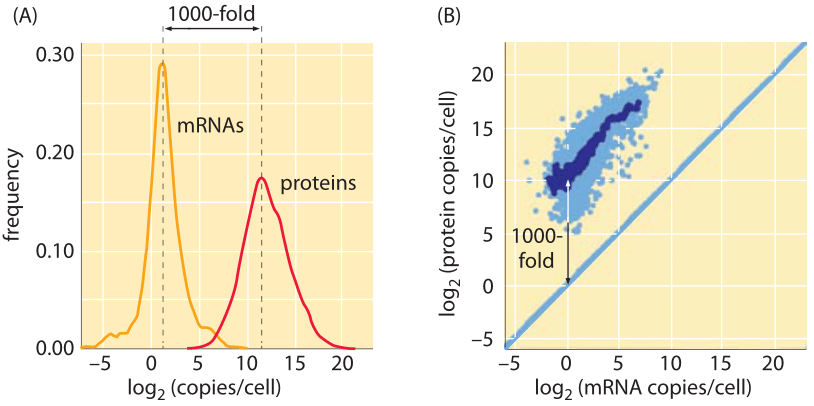
Figure 3: Protein to mRNA ratio in fission yeast. (A) Histogram illustrating the number of mRNA and protein copies as determined using sequencing methods and mass spectrometry, respectively. (B) Plot of protein abundance and mRNA abundance on a gene-by-gene basis. Adapted from S. Marguerat et al., Cell, 151:671, 2012. Recent analysis (R. Milo, Bioessays, 35:1050, 2014) suggests that the protein levels have been underestimated and a correction factor of about 5-fold increase should be applied, thus making the ratio of protein to mRNA closer to 104.
As with many of the quantities described throughout the book, the high-throughput, genome-wide craze has hit the subject of this vignette as well. Specifically, using a combination of RNA-Seq to determine the mRNA copy numbers and mass spectrometry methods and ribosomal profiling to infer the protein content of cells, it is possible to go beyond the specific gene-by-gene estimates and measurements described above. As shown in Figure 3 for fission yeast, the genome-wide distribution of mRNA and protein confirms the estimates provided above showing more than a thousand-fold excess of protein to mRNA in most cases. Similarly, in mammalian cell lines a protein to mRNA ratio of about 104 is inferred (BNID 110236).
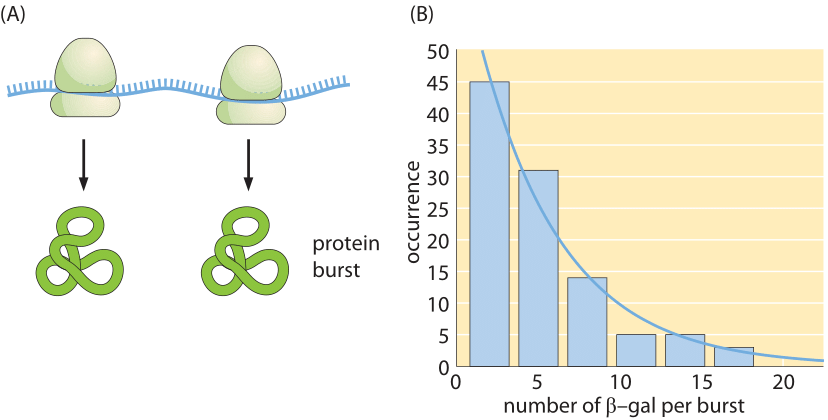
Figure 4: Dynamics of protein production. (A) Bursts in protein production resulting from multiple rounds of translation on the same mRNA molecule before it decays. (B) Distribution of burst sizes for the protein beta-galactosidase in E. coli. (Adapted from L. Cai et al., Nature, 440:358, 2006.)
So far, we have focused on the total number of protein copies per mRNA and not the number of proteins produced per production burst occurring from a given mRNA. This so-called burst size measurement is depicted in Figure 4, showing for the protein beta-galactosidase in E. coli the distribution of observed burst sizes, quickly decreasing from the common handful to much fewer cases of more than 10.
Finally, we note that there is a third meaning to the question that entitles this vignette, where we could ask how many proteins are made from each individual mRNA before it is degraded. For example, in fast growing E. coli, mRNAs are degraded roughly every 3 minutes as discussed in the vignette on “What is the degradation rates of mRNA and proteins?”. This time scale is some 10-100 times shorter than the cell cycle time. As a result, to move from the statement that the protein to mRNA ratio is typically 1000 to the number of proteins produced from an mRNA before it is degraded we need to divide the number of mRNA lifetimes per cell cycle. We find that in this rapidly dividing E. coli scenario, each mRNA gives rise to about 10-100 proteins before being degraded.
A recent study (G. Csardi et al., PLOS genetics, 2015) suggests revisiting the basic question of this vignette. Careful analysis of tens of studies on mRNA and protein levels in budding yeast, the most common model organism for such studies, suggests a non-linear relation where genes with high mRNA levels will have a higher protein to mRNA ratio than lowly expressed mRNAs. This suggests the correlation between mRNA and protein does not have a slope of 1 in log-log scale but rather a slope of about 1.6 which also explains why the dynamic range of proteins is significantly bigger than that of mRNA.

