Information & Errors – Introduction
What is it that makes living matter so different from its inanimate counterpart? Stated simply, living matter carries within it the blueprint for its own construction. The storehouse of information contained both in genomes and in the post-translational modifications of proteins leads to an ability to pass information along from one generation to the next with staggering fidelity. Genomes preside over the management of the molecules of the cell in ways that forbid them from becoming an inactive soup of chemicals whose potential for further reactions has been exhausted. This feat is all the more impressive given that on evolutionary time scales, this information content changes as a result of adaptations and genetic drift.
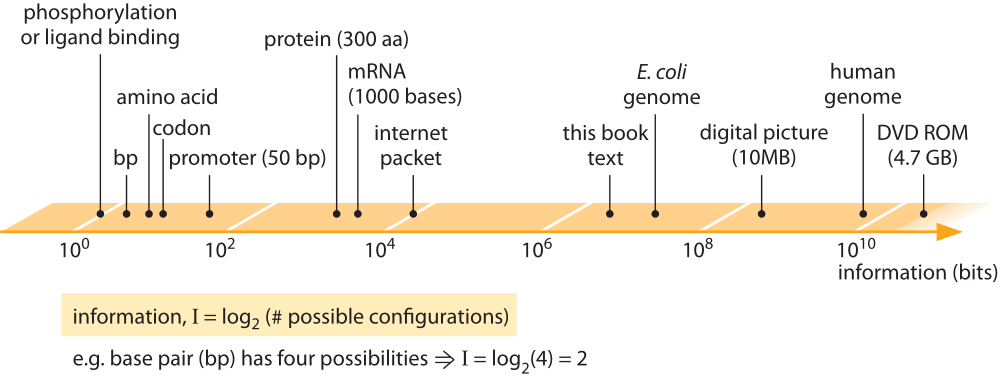
Figure 1: Information content of biological entities and some man made information storage devices. Information is quantified through binary bits, where a base pair which has 4 possibilities is 2 bits etc.
The vignettes presented in this chapter all focus either directly or indirectly on quantifying the management of the information content in cells. The scale of information storage in biological components is depicted in Figure 1 and compared to man-made information storage devices. The juxtaposition of biological and human information storage is both surprising and enlightening. To get a sense of the astonishing information density of biological systems, consider an estimate made by one of our students in a class on “Cell Biology by the Numbers’’. What this student found is that if one imagines the information storage density of the influenza virus scaled up to the size of a modern disk-on-key device it would account for several exabytes of data (1018), equivalent to the global internet traffic over a few days.
In this chapter, we begin by examining genomes themselves. How big are they and how many genes do they harbor? We will see that there is a huge eight orders of magnitude (or even more) difference in sizes between the largest and smallest genomes, though the number of genes they contain shows much less variability. The next set of questions that we broach in considering information management within cells center broadly on the question of biological fidelity. In the Middle Ages, the promulgation of sacred texts took place through the patient action of scribes whose job it was to copy the contents of these books. Like any copying process, these reproductions were subject to mistakes and it is the biological analog of such mistakes that will concern us here. We mainly examine the error rate associated with the processes of the central dogma. How many mistakes are made each time a genome is copied? When new proteins are synthesized, how often is the wrong amino acid added onto the nascent polypeptide chain?
We then expand the scope of our discussion to ask about substrate recognition more broadly. Many proteins have their activity shifted by the addition and removal of charged groups such as phosphates through the action of kinases. But what prevents these kinases from adding a phosphate group on the wrong substrate and how often are such mistakes made? After all, in general, kinases add phosphates to only a very limited set of amino acids which are shared by nearly all proteins and hence, it is of great interest to better understand the discriminatory powers that are exploited in selecting residues for phosphorylation.
All told, information management is one of the great themes of biology and the task of this chapter is to provide a quantitative view of some of these questions.

