What is the turnover time of metabolites?
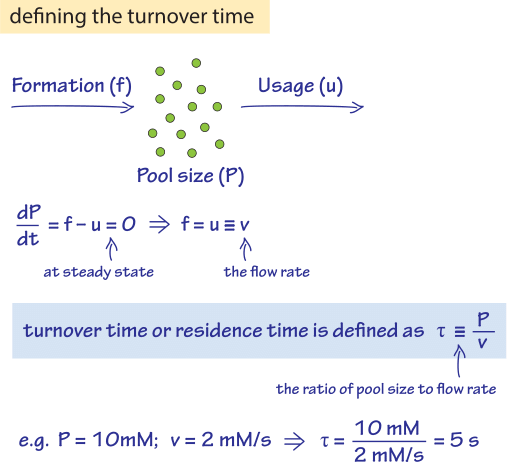
Figure 1: The turnover time is defined through the ratio of the pool size to the flux. In steady state the flux is equal to the formation rate which also equals the usage rate.
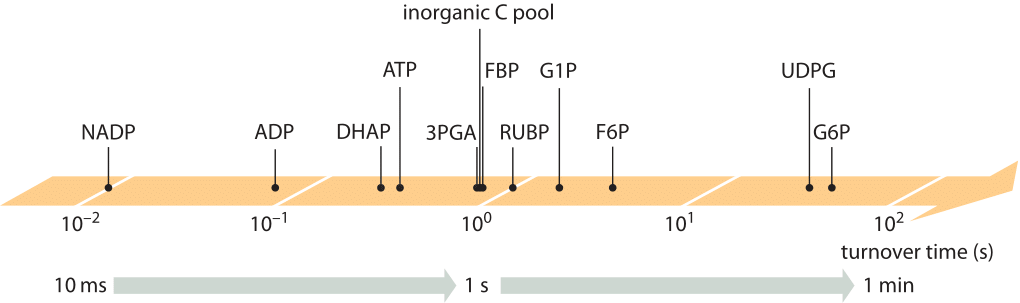
Fast cellular growth rates are associated with proportionally higher utilization rates (fluxes) of precursor metabolites. At the same time the concentrations of intermediate metabolites need to be kept at levels not exceeding a few mM, to avoid problems ranging from osmotic pressure imbalance to non-specific cross reactivity. Achieving these two aims, namely high fluxes at low intermediate concentrations, implies a quick turnover time of the metabolite pool. The turnover time concept is schematically shown in Figure 1 and is defined to be the mean time over which the pool of a given metabolite will be replaced due to the rates of production and utilization (which are equal in steady state). Indeed for many key metabolites of central carbon metabolism the turnover time is on the order of a second as shown in Figure 2 and Table 1 for the case of the model plant Arabidopsis Thaliana (BNID 107358). Similarly, in E. coli, the pools of most amino acids were shown to turnover in less than a minute (BNID 101622). The subsecond turnover times in Arabidopsis manifest in the startling finding that when aiming to perform a metabolomics experiment that measures the concentration of metabolites, if the researcher briefly passes a hand over the light source when heading to throw the plant into the liquid nitrogen, the result will already be different for Calvin-Benson cycle metabolites than if the researcher was careful not to block the light.
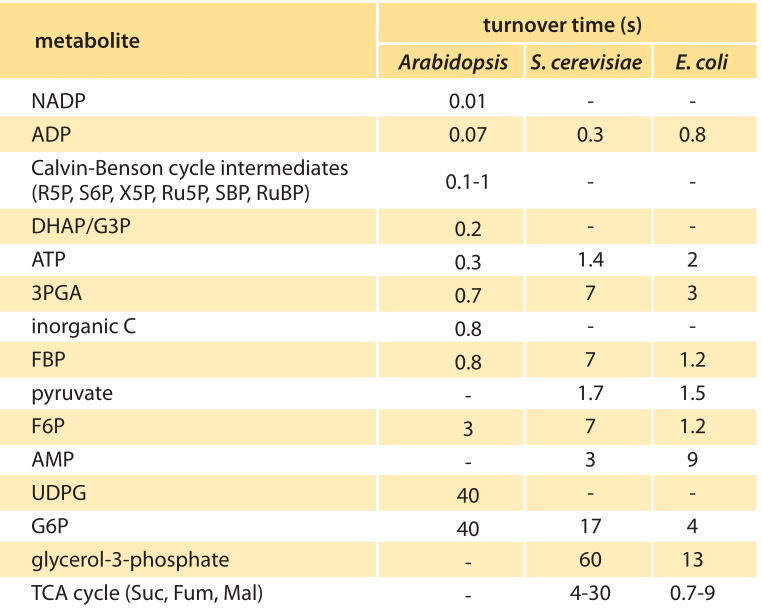
Table 1. Metabolite turnover times in an Arabidopsis circular arrangement of leaves (rosette) were measured by mass spectrometry. Turnover times for metabolites in the Calvin cycle, and starch and sucrose synthesis, under light and 485 ppm CO2 were calculated. Adapted from Arrivault et al., Plant J., 2009. Data for E. coli and S. cerevisiae are from BNID 109701.
Protein synthesis provides another example of fast turnover, where a high rate of polymerization of monomeric amino acids takes place mediated by shuttling by tRNAs. Yet, the total number of tRNAs is limited. In E. coli growing with a doubling time of 40 minutes the total number of tRNAs is estimated at ≈200,000 copies per cell (BNID 100066). Given that there are about 30,000 ribosomes (BNID 102015) each working at a rate of polymerization of ≈20 aa per second (BNID 100059) the average turnover time is about 200,000/(20 s-1 x 30,000)≈1/3 of a second. This is the time frame between loading an amino acid onto the tRNA through tRNA synthetase, binding of that tRNA within a ribosome where the amino acid is released and forms the peptide bond, and the replenishment of that tRNA by the loading of a new amino acid. Though this estimate has been carried out in less than one paragraph, careful experiments to actually obtain precise measurements were much harder. Using radioactive pulse-labeling the numbers from the estimate above were confirmed, resulting in a range of turnover times between 0.1-1 second (BNID 105275) for the turnover of the tRNA pool. In budding yeast the corresponding numbers are about 2 million tRNAs (BNID 108197), 200,000 ribosomes (BNID 100267, 108197) and a polymerization rate of ≈10 aa per second (BNID 107785, 107871) resulting yet again at a turnover time of about 1 s. As an aside, we note that the ratio of total tRNA per ribosome tends to be about 10 to 1.
The variety of surprising numbers for turnover times offered throughout this vignette paint a vibrant picture of the chemical hustle and bustle taking place within cells. Though many of our structural descriptions of biology offer a static picture of the molecules of the cell, we see here that whether talking about the molecular components of central metabolic pathways or key players in the central dogma such as the tRNAs that make protein synthesis possible, these molecules often are transitioning between different states literally in the blink of an eye (taking about 0.1-0.4 s, BNID 100706).