Which is bigger, mRNA or the protein it codes for?
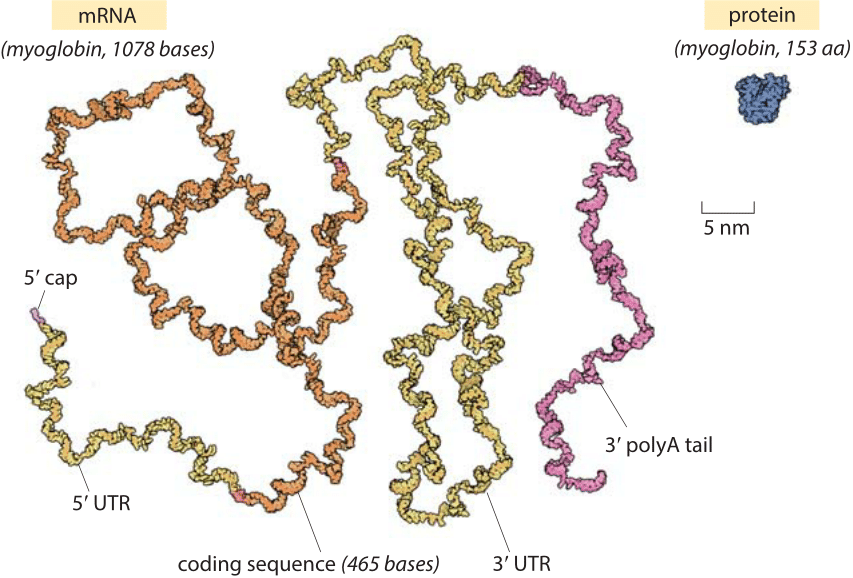
Figure 1: The relative sizes of a globular protein and the mRNA that codes for it. The myoglobin protein is drawn to scale next to the mRNA transcript that leads to it. The coding sequence of an mRNA alone is about an order of magnitude heavier by mass than the protein. The myoglobin protein is in blue, the 5’ cap and 3’ polyA tail are in purple, the 5’ and 3’ untranslated regions (UTRs) are in yellow and the coding sequence is in orange. Illustration by David Goodsell.
The role of mRNAs as epitomized in the central dogma is one of fleeting messages for the creation of the main movers and shakers of the cell, namely, the proteins that drive cellular life. Words like these can conjure a mental picture in which an mRNA is thought of as a small blueprint for the creation of a much larger protein machine. In reality, the scales are exactly the opposite of what most people would guess. Nucleotides, the monomers making up an RNA molecule, have a mass of about 330 Da (BNID 103828). This is about 3 times heavier than the average amino acid mass which weighs in at ≈110 Da (BNID 104877). Moreover, since it takes three nucleotides to code for a single amino acid, this implies an extra factor of three in favor of mRNA such that the mRNA coding a given protein will be almost an order of magnitude heavier when one compares codons to the residues they code for. A realistic depiction of a mature mRNA versus the protein it codes for, in this case the oxygen-binding protein myoglobin, is shown in Figure 1. As can be clearly seen in the figure, in the microscopic world, our everyday intuition that the blueprint (mRNA) should be smaller than the object it describes (protein) does not hold. In eukaryotes, newly transcribed mRNA precursors are often richly decorated with introns that skew the mass imbalance even further.

Figure 2: Cryo-electron microscopy images of RNAs in vitrified solution. (A) Fourier band-pass filtered images of a 975-nt RNA from chromosome XII of S. cerevisiae. Individual RNA molecules, suspended in random orientations, are seen as dark branched objects. (B) Traced skeletons of the molecules in panel A. (C) depiction of a hundred traced projections superimposed with their centers of mass in registry. (B adapted from A. Gopal et al., RNA 18:284, 2012.)
What about the spatial extent of these mRNAs in comparison with the proteins they code for? the mass excess implies a larger spatial scale as well, though the class of shapes adopted by RNAs are quite different than their protein counterparts. Many proteins are known for their globular structures (see vignette “How big is the “average” protein?”). By way of contrast, mRNA is more likely to have a linear structure punctuated by secondary structures in the form of hairpin stem-loops and pseudoknots, but is generally much more diffuse and extended. The “thread-like” mRNA backbone has a diameter of less than 2 nm, much smaller than the diameter of a characteristic globular protein of about 5nm (BNID 100481). On the other hand, a characteristic 1000 nucleotide long mRNA (BNID 100022) will have a linear length of about ≈300 nm (BNID 100023). The most naïve estimate of mRNA size is to simply assume that the structure is perfectly base paired into a double stranded RNA molecule. For a 1000 base long mRNA, this means that its double-stranded version will be 500 bp long, corresponding to a physical dimension of more than 150 nm, using the rule of thumb that a base pair is about 1/3 nm in length. This is an overestimate since these structures are riddled with branches and internal loops which will shorten the overall linear dimension. Recent advances have made it possible to visualize large RNA molecules in solution using small angle X-ray scattering and cryo EM as shown in Figure 2. One of the useful statistical measures of the spatial extent of such structures is the so-called radius of gyration which can be thought of as the radius of a sphere of an equal effective size. For RNAs this was found to be roughly ≈20 nm (BNID 107712) indicating a characteristic diameter of ≈40 nm. Hence, contrary to the expectation of our uncoached intuition, we note that like the mass ratio, the spatial extent of the characteristic mRNA is about 10 fold larger than the characteristic globular protein.

